Corresponding Author: Linda J. Mullins
Queen’s Medical Research Institute, Cardiovascular Science Centre,
University of Edinburgh, Little France Crescent, Edinburgh, EH16 4TJ (UK)
Tel. (44) 0131 242 6720 , E-Mail linda.mullins@ed.ac.uk
scRNA Transcription Profile of Adult Zebrafish Podocytes Using a Novel
Reporter Strain
Cara Browna Linda J. Mullinsa Katrina Wesencraftb Gail McConnellb Mariana Beltranc Neil C. Hendersonc,d Bryan Conwaya Scott Hoffmanna Sebastian Ridera,e John J. Mullinsa
aQueen’s Medical Research Institute, Cardiovascular Science Centre, University of Edinburgh, Edinburgh, UK, bDepartment of Physics, SUPA, University of Strathclyde, Glasgow, UK, cQueen’s Medical Research Institute, Centre for Inflammation Research, University of Edinburgh, Edinburgh, UK, dMRC Human Genetics Unit, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK, eDSM Nutritional Products FranceCRNA, Village-Neuf, France
Introduction
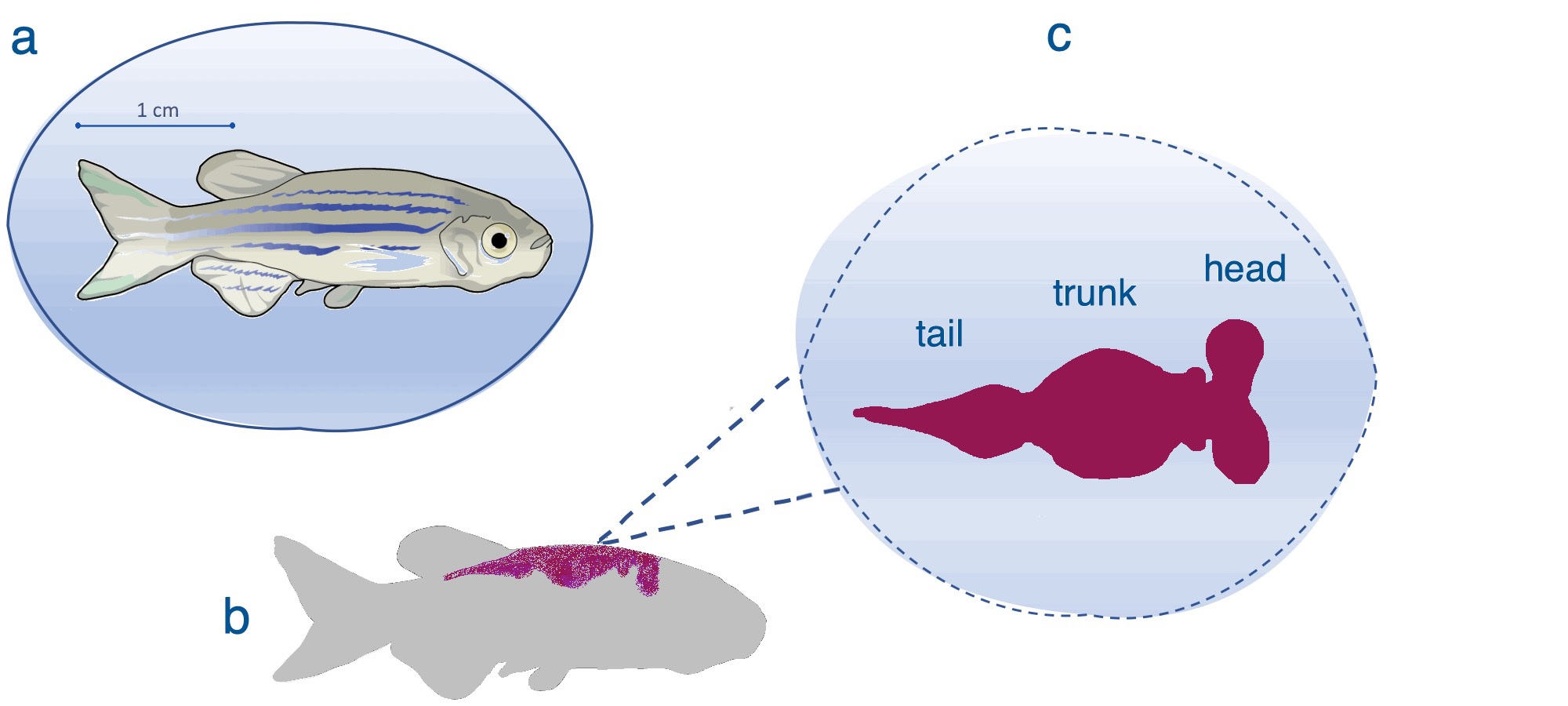
Located within the glomerular capsule, the podocyte forms the outermost layer of the glomerular filtration barrier - a key component of the nephron, which is the functional unit of the kidney (Fig. 1). The fundamental importance of podocytes is highlighted by the conserved nature of podocyte function across a myriad of species [1-3]. Podocytes play a vital role in filtration, forming a selectively permeable barrier, preventing larger charged proteins within the glomerular capillaries from passing through into the urinary ultrafiltrate in the capsular space [4].
Podocytes are highly ramified epithelial cells, which envelope the glomerular capillaries (Fig. 1d, e). The podocyte is composed of three main cellular structures, the cell body, the main processes and the foot processes (pedicels) that form finger-like projections [5].
These interdigitate with foot processes of neighbouring podocytes, leaving minute gaps between them. These gaps, known as slit diaphragms, are a specialised type of intercellular junction, the formation of which, requires nephrin, and the podocyte-specific podocin, amongst other proteins [6-8].
Podocin is pivotal to maintaining both the development of lipid rafts and the structural integrity of the slit diaphragm [9].
Podocytes are often referred to as a type of specialised renin precursor cell. Podocytes can communicate with one another using neurone-like signalling through the release of neurotransmitter glutamate, which is released via vesicles comparable to the glutamatergic signalling system seen in neurones [10]. Calcium ions also play an essential part in podocyte cell interaction and signalling [11].
Podocyte damage is a common pathology observed in renal disease, such as Focal Segmental Glomerulosclerosis [12, 13] or diabetic nephropathy [14] - evidence of their vital role in kidney functionality. Cell function can be adversely affected by exposure to toxins, which disrupt the organisation of actin filaments within the cells, and lead to podocyte effacement [15, 16] and proteinuria. The zebrafish is becoming a key model for in vivo drug screening [17] during larval kidney development, and adult kidney injury [18, 19].
The transgenic (tg) line Tg(-2.5nphs2:GCaMP6s,P2A,LifeAct-TagRFP-T) was designed to specifically express two reporters under the podocin promoter - the calcium indicator, GCaMP6s [20, 21], which therefore potentially marks early developing or damaged podocytes [22], together with LifeAct-TagRFP-T, a red fluorescence protein conjoined with LifeAct, which binds to F-actin [23]. This allows visualisation of the actin filaments that are intimately involved in the cytoskeletal architecture of the podocyte foot processes [24]. Adult transgenic zebrafish were used as the source of podocytes, which were FAC sorted and processed for single cell RNA sequencing (scRNA-seq), to provide their transcriptional profile. This was compared with the profiles from higher organisms.
Animal Husbandry
All experiments were conducted and approved by the University of Edinburgh Animal Welfare and Ethical Review Body (AWERB) and in accordance with the UK Home Office Animals Scientific Procedures Act 1986. Zebrafish (Danio rerio; WIK background) were maintained at 28.5°C [25]. Adult fish were anaesthetized via immersion in 4.2 mg/ml tricaine and the kidney was harvested at this point.
Generation of Fish Lines
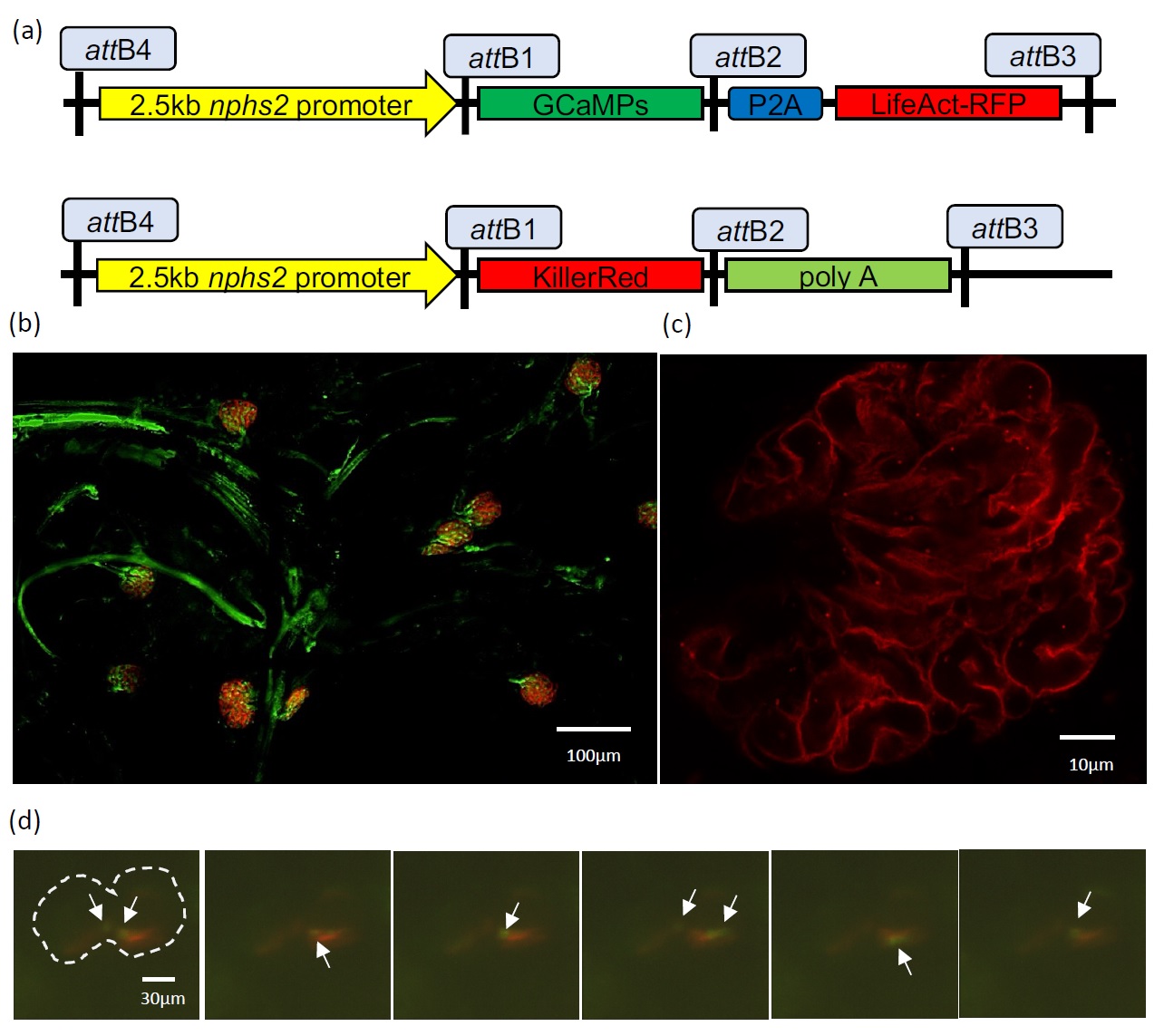
The transgenic zebrafish lines used in this work were Tg(-2.5nphs2:GCaMP6s,P2A,LifeAct-TagRFP-T;cryaa:CFP) and a cross between Tg(-2.5nphs2:KillerRed;cryaa:CFP) and Tg(flk:EGFP). The transgene constructs were created using gateway cloning (Invitrogen) in an expression vector carrying the cyan reporter under the eye-specific crystallin, alpha a promoter (cryaa:CFP). The component parts of the expression vector (Fig. 2a) were a 2.5-kb podocin (nphs2) promoter, which had previously been shown to direct podocyte-specific expression in mice [26] driving either the GCaMP6s calcium indicator linked through a P2A cleavage site to the LifeAct-TagRFP-T or KillerRed. Expression vectors were co-injected with transposase mRNA, using the Tol2 kit (Invitrogen), into wild type WIK zebrafish embryos.
Microscopy
Incisions were made either side of the abdomen, and skin, swim bladder and internal organs were peeled back to expose the kidney. The kidney (Fig. 1b-c) was peeled away from the backbone, placed under a cover slip with a drop of phosphate buffered saline, gently compressed to flatten the sample, sealed and mounted on the DMI8-CS Sp8 confocal microscope. Images were obtained using a 63x/ 1.4NA oil HC PL APO Cs2 objective with ALEXA 488 and mCherry filters.
Mesolens
The Mesolens is a giant objective lens with the unique combination of low magnification and high numerical aperture (4x/0.47) [27]. The lens is designed for confocal imaging of tissue volumes up to 100 mm3 with sub-cellular resolution throughout. Specimens were mounted in phosphate buffered saline (Thermo Fisher, GIBCO) within a custom mounting chamber for long-term immersion [28]. Two laser lines with wavelengths of 488nm and 561nm (Omicron) at average powers of 3mW and 12mW respectively were used for simultaneous excitation of fluorescence from EGFP and KillerRed. The fluorescence signals were separated using a 550nm long-pass dichroic filter (DMLP550R Thorlabs), with the EGFP signal propagating through a 525/39 bandpass filter (MF525-39) and the KillerRed signal passing through a 600nm long-pass filter (FEL0600, Thorlabs). The fluorescence signals were detected by individual photomultiplier tubes. Images were acquired with Nyquist sampling in all dimensions (pixel size of 500nm in xy, z-step size of 3 µm), and each optical section was averaged over n=2 frames. Images were deconvolved with Huygens Professional version 19.04 (Scientific Volume Imaging, http://svi.nl), using the CMLE algorithm, with a signal-to-noise ratio of 40:1 and 10 iterations. Digital movies zooming into regions of interest were created using the FIJI distribution of ImageJ [29].
Selective Plane Illumination Microscopy (SPIM)
Healthy embryos from the Tg(-2.5nphs2:GCaMP6s,P2A,LifeAct-TagRFP-T;cryaa:CFP) line were treated with 1-phenyl-2-thiourea (PTU) at approximately 8 hpf (hours post fertilisation) to suppress the development of pigmentation. From 3 dpf (days post fertilisation), larvae were selected and immersed in mivacurium chloride at 0.5 mg/ml. After 10 minutes, the immobilised larvae were mounted individually in 1% low melting point agarose in a capillary tube attached to a syringe. The capillary tube’s open end was capped and undisturbed until the agarose had solidified, holding the larvae in place. The apparatus containing the syringe, the capillary tube and the immobilised larva was mounted into our custom built SPIM microscope [30] for imaging. The larva was orientated so that the glomerulus was visible. Images were obtained using a 16X 0.8 NA Nikon CFI LWD Plan Fluor water dipping objective (N16LWD-PF). Laser excitation was at 488 and 561 nm, using a Versalase laser system (Vortran), as previously published [31].
Laser power of 11mW was used to orientate the larva without induction of a calcium response. Laser power of 20mW induced injury in podocytes, and the subsequent presence of Ca2+ ions was detected by the calcium sensor, GCaMP6s, within the podocytes. The resultant green fluorescence was recorded over time (acquisition time 0.2s; interval between images 15s). After imaging, the larva was removed from the capillary tube and allowed to recover briefly in conditioned water at 28.5°C and was then fixed for later analysis.
Dissociation of Podocytes from the Kidney
Adult Tg(-2.5nphs2:GCaMP6s,P2A,LifeAct-TagRFP-T;cryaa:CFP) zebrafish were used as the source of podocytes. Kidneys were dissected as above and transferred to ice-cold Leibowitz-15 (L-15) medium. Cells were dissociated in medium supplemented with the psychrophilic enzyme, cold activated protease (NATE0633, 20mg/ml), DNAse (400units/ml), Liberase (NATE1785; 85µg/ml) and collagenase IV (2mg/ml) and CaCl2 (5mM), at 6°C for 15 minutes with trituration every 5 minutes. The temperature was increased to 28°C for a further 15 minutes, (again with trituration, but using a smaller diameter pipette tip). Dissociated kidneys were passed through a 40 µm cell strainer before centrifugation. Cells were resuspended prior to FAC sorting in PBS with 2% fetal calf serum. A WIK mesonephric kidney was used as an auto-fluorescent control (blue excitation 488nm; emission filter 695/40). Live-dead cell count was assessed by 4′,6-diamidino-2-phenylindole stain ( DAPI-UV excitation 360nm; emission filter 450/50) and singlets (FSC-A versus SSC-A) were gated for red fluorescence (excitation 561nm; emission filter 582nm/15-A), using the BD FACS Aria II SORP (Becton Dickinson, Basel, Switzerland) with a 100µm nozzle.
10x Chromium single cell Library workflow
Single cells were processed using the Chromium™ Single Cell 3′ Library and Gel Bead Kit v2 (10X Genomics, PN-120237) and the Chromium™ Single Cell A Chip Kit (10X Genomics, PN-120236) as per the manufacturer’s instructions. In brief, single cells were sorted into PBS + 2% FBS, and washed once. An estimated 7-10,000 cells were added to each lane of a 10X chip and partitioned into Gel Beads in emulsion, where cell lysis and barcoded reverse transcription of RNA occurred, followed by amplification, fragmentation and 5′ adaptor and sample index attachment. Libraries were sequenced on an Illumina HiSeq 4000.
Transcriptome libraries were mapped to a Danio rerio reference genome constructed from the zebrafish GRCz11 genome assembly. Briefly, reporter gene coding sequences were fused to the Ensembl 96 gtf file and the reference genome was built using the cellranger mkref software. Single cell RNA sequences, with associated UMIs, were aligned to this amended reference genome using Cell Ranger v2.1.0 Single-Cell Software Suite from 10X Genomics.
The resultant datasets were analysed using the Seurat R package v2.4.3 [32] as per the clustering workflow. Briefly, genes expressed in fewer than three cells or cells expressing fewer than 200 genes or mitochondrial gene content > 30% of the total UMI count were excluded. We normalized using the global-scaling “LogNormalize” transformation. Highly variable genes were identified using Seurat’s ‘FindVariableGenes’ function with default parameters. Dimensionality was reduced by principal component analysis (PCA). We performed unsupervised clustering and differential gene expression analyses using SNN graph-based clustering, and the first 18 principal components as determined by variability in the PC Elbow Plot. The number of clusters was tuned using the resolution parameter. Heatmaps, t-SNE visualizations, and violin plots were produced using Seurat functions. Pseudotime between clusters was assessed using the Monocle workflow in R [33].
Results
As part of a wider strategy to develop tools for understanding mechanisms underlying renal damage, we established a series of novel transgenic zebrafish strains, expressing fluorescent reporters specifically within podocytes. These include strains with the podocin promoter driving expression of LifeAct-RFP, in which the fluorophore binds to the cytoskeleton; GCaMP6s, which binds to Ca2+; and KillerRed, through which optogenetic approaches allow cell specific ablation of podocytes [30]. These strains were crossed with lines expressing fluorescent reporters marking vasculature or kidney tubules, as required. Representative images, showing fluorescent podocytes expressing KillerRed or LifeActRFP within the fish glomeruli, captured using the Mesolens and confocal microscopy respectively, are shown (Fig. 2b, c; Supplementary Movie 1 – for all supplementary material see www.cellphysiolbiochem.com).
The LifeAct-RFP fluorescent reporter faithfully replicated podocin expression (Fig. 2c) and was restricted to podocytes in both the pronephros and mesonephros (only the latter is shown), during imaging by light sheet microscopy. We discovered that increasing laser power to 20mW caused spontaneous podocyte injury in pronephric glomeruli and fluorescence of the calcium sensor, suggesting Ca2+ uptake/release into the podocyte cytoplasm. Green fluorescence was seen at multiple locations over the course of the experiment, and exemplary stills are shown (Fig. 2d; full video: Supplementary Movie 2).
Cell dissociation and FACs sorting
Zebrafish kidneys were initially treated with a cocktail of enzymes, including cold activated protease, at 6°C. This dissociated the kidney sufficiently to release glomeruli, which were dissociated further by transferring the mixture to 28°C. The two-stage dissociation protocol minimised exposure of podocytes to a higher temperature and thus limited cell damage prior to FAC sorting (for typical run see Supplementary Fig. S1). Eight adult kidneys yielded sufficient numbers of podocytes for the generation of a 10X scRNA-seq library. Cell Ranger v2.1.0 Single-Cell Software Suite yielded information on approximately 2,200 cells.
Identification of Cell Clusters
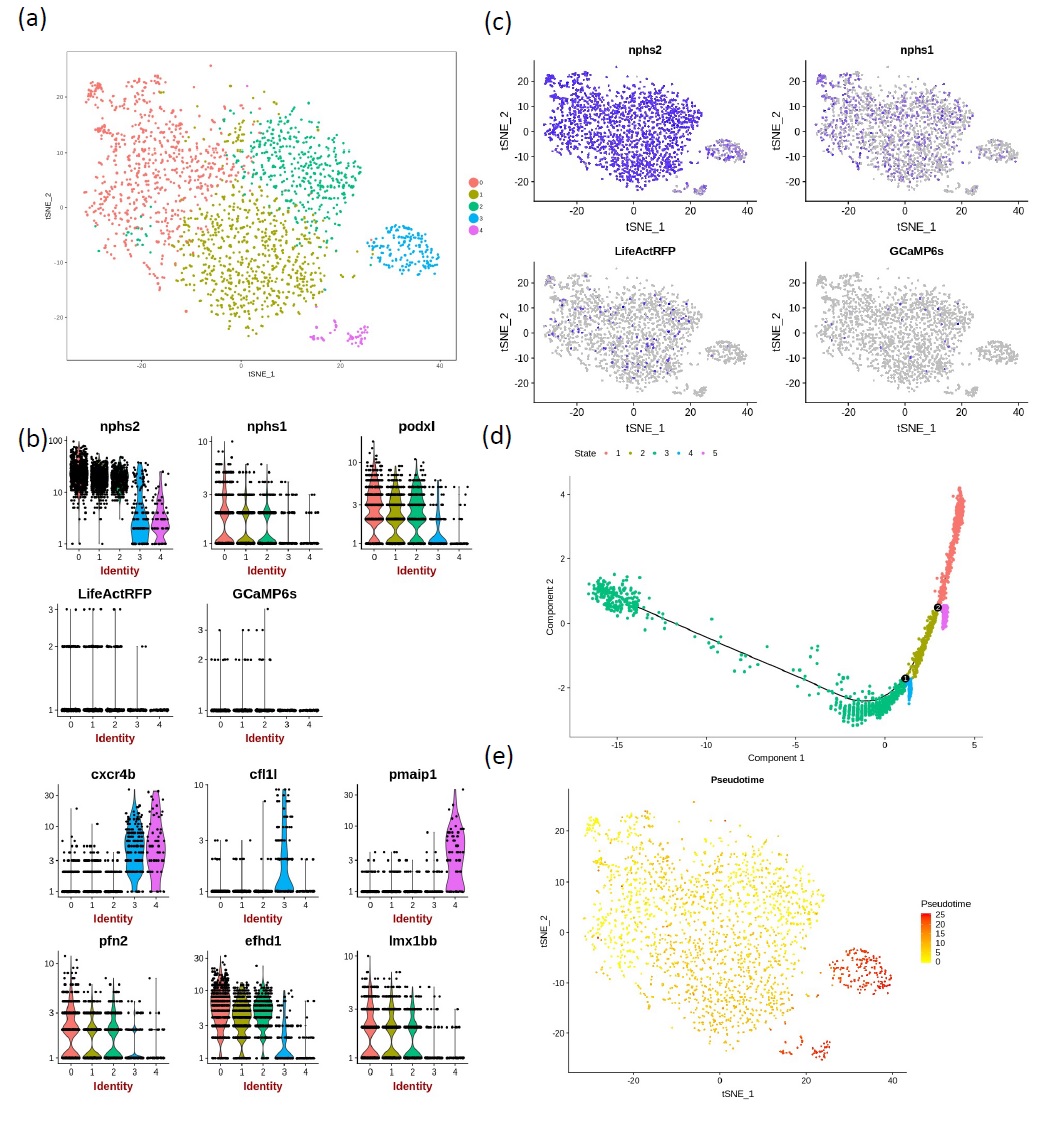
Principal component analysis allowed non-linear dimensional reduction of the scRNA-seq data and tSNE plots were used to group similar cells into clusters, the number of which was adjusted by altering the resolution. Increase in the resolution parameter sets the granularity of clustering leading to higher numbers of clusters. Since the mesonephric kidney grows continuously in the zebrafish, we wanted to separate podocytes spanning a range of stages in development and maturity. Resolution 0.3 returned five clusters of cells (Fig. 3a) and was used in all further analysis.
Violin plots were used to show the expression probability distributions across clusters (Fig. 3b). These plots show that expression of podocin (nphs2), nephrin (nphs1) and podocalyxin-like (podxl) was observed across all clusters, and was highest in groups 0, 1 and 2. Likewise, the two reporters LifeActRFP and GCaMP6s were most highly expressed in clusters 0, 1 and 2. The overlap of gene expression across clusters could be attributed to the up or down-regulation of specific genes, during podocyte maturation. Other genes showing a similar distribution were profilin 2 (pfn2), which positively promotes actin filament assembly, the calcium-binding protein gene, efhd1, and the transcription factor lm1bb. Genes which showed increased expression in clusters 3 and/or 4 included the cytokine receptor cxcr4b, cofilin-1-like (cfl1l), which is involved in actin filament depolymerization and the apoptotic regulator, pmaip1.
Feature plots were used as an alternative way to demonstrate the extent of podocin and nephrin expression across the clusters in comparison to the expression of the fluorescent reporter genes (Fig 3c). These again show that podocin is highly expressed across podocytes, while GCaMP6s and LifeActRFP show lower levels of steady state transcription under the podocin promoter. It should be noted that cells were sorted using RFP fluorescence, and that podocin is a marker of mature podocytes. The scRNA-seq data provide a snapshot of the genes that were actively transcribed at the time of processing. Although the fluorescent protein was present in the cell during FAC sorting, this plot suggests that the GCaMP6s and LifeAct-RFP genes were not very actively transcribed, or that the transcripts are less stable than the endogenous podocin transcript (Fig. 3b, c). scRNAseq data is deposited at Edinburgh Datashare (https://doi.org/10.7488/ds/3021).
Comparisons with Gene Expression in alternative species
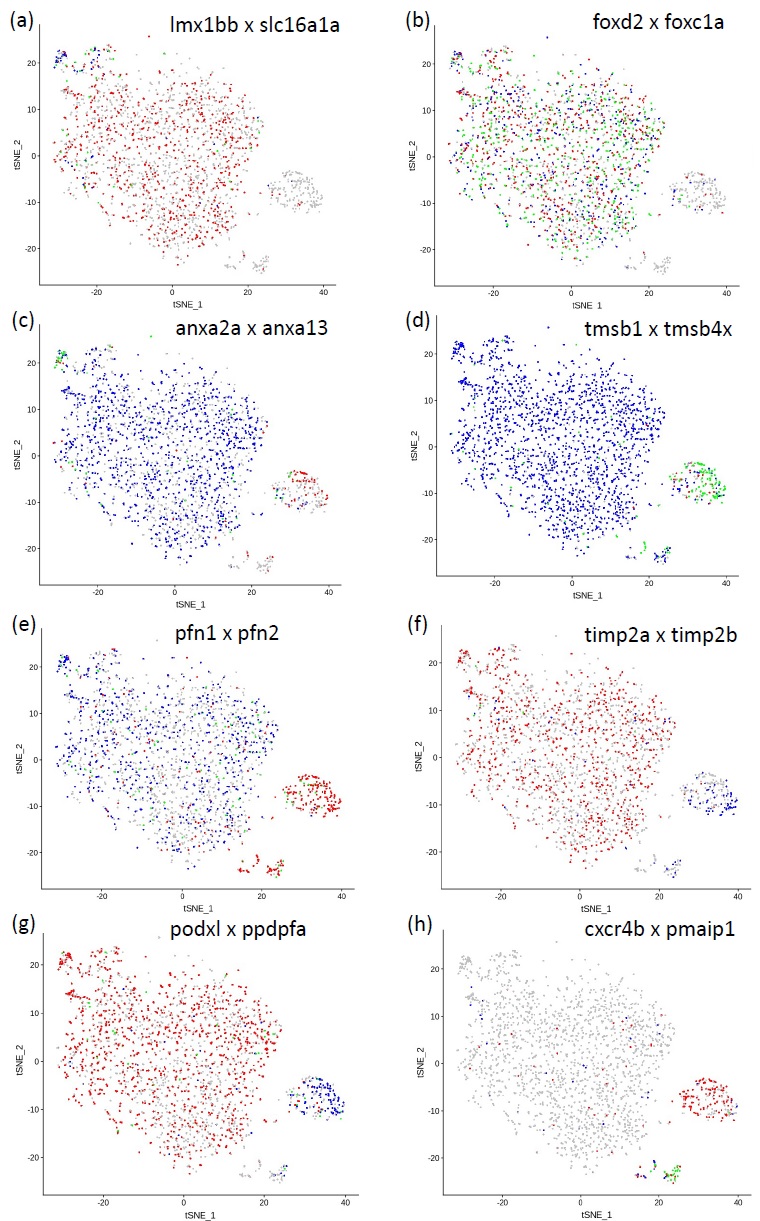
The gene transcription profile of podocytes identified in our zebrafish scRNA-seq dataset was compared with corresponding transcription profiles from human [34] and mouse [35, 36] podocytes. We found very few markers designated ‘early’ from human data apart from the monocarboxylate transporter, slc16a1a, which was transcribed in a small subset of cluster 0 cells (Fig. 4a) and the ‘early’ transcription factor, lmx1bb, though this was more widely distributed through clusters 0, 1 and 2, suggesting it is retained in more mature podocytes. Additional transcription factors identified were foxd2 and foxc1a
(Fig. 4b) and mafba (data not shown). This probably reflects the fact that we FAC sorted podocin-expressing cells, and podocin is not expressed very early in podocyte development.
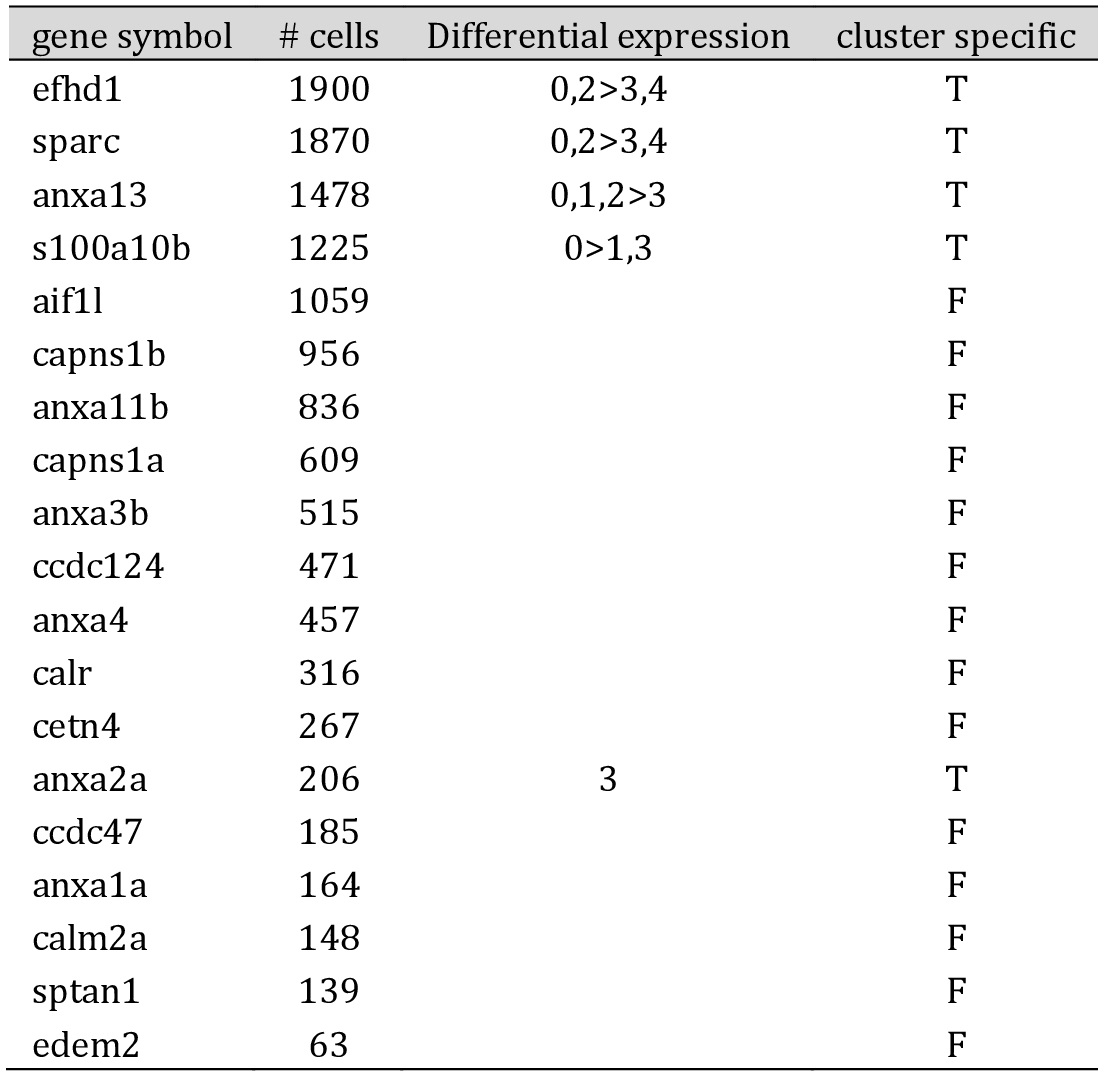
Genes expressing calcium-ion binding activity are associated with podocyte development, and the presence of these genes in a cluster may indicate that it contains young or developing podocytes. The zebrafish scRNA-seq data identified a number of genes including osteonectin (sparc), efhd1, annexins 2a and 13 (anxa2a; anxa13) and S100A10b, which were differentially expressed between the clusters (Table 1). Anxa13 was widely expressed but anxa2a was limited to podocytes also expressing slc16a1a (proposed early podocytes) or those in clusters 3 (see Fig. 4c)
Genes involved with actin filament binding or assembly are shown in Table 2. These included myozenin 1b (myoz1b), syndecan 4 (sdc4), myosin light chain 9a (myl9a), profilins 1 and 2 (pfn1; pfn2) and thymosins b1 and b4x (tmsb1; tmsb4x), many of which were differentially expressed. Of note, tmsb1 and pfn1 were limited to clusters 3 and 4 (Fig. 4c, d).
The other genes were expressed extensively throughout clusters 0, 1 and 2, and are associated with developing or mature podocytes.
A large number of ‘late’ podocyte markers [34] were found in the zebrafish dataset, including vegfaa and vegfab, col4a3 and col4a4, clic2, gadd45a and gsna, all of which were widely expressed, as was connexin 43, encoded by cx43, which contributes to gap junctions, providing routes of intracellular diffusion. Of interest, were a number of genes differentially expressed between clusters 0, 1 and 2 and clusters 3 and 4 (Fig. 4).
Podocalyxin-like (podxl), and the transcription factor mafba are both involved in pronephric glomerular morphogenesis, and were expressed extensively. However, ppdpfa expression, which is related to cell fate, and cfl1l, which is involved in the regulation of cytoskeletal dynamics and acts to depolymerise filamentous actin, were found in cluster 3, while pmaip1, along with the chemo-attractant receptor, cxcr4b, were distributed in clusters 3 and 4. This suggested that clusters 3 and 4 contain more mature podocytes initiating apoptosis and the subsequent signalling of clearance by phagocytes [37, 38].

Pseudotime analysis
Pseudotime analysis using Monocle suggested that there were five distinct states in the cell expression data, indicating a progression through stages in podocyte development (Fig. 3d). We were able to superimpose pseudotime on the Seurat-derived clusters (Fig. 3e), suggesting that clusters 3 and 4 are at a later stage in development. This confirms our conclusion that these clusters may represent aging podocytes earmarked for disposal. It should be noted that changes in transcription levels do not necessarily translate to changes in protein levels.
Discussion
The new transgenic zebrafish lines we describe here should prove useful in the understanding of podocyte development and injury processes. We have demonstrated that both KillerRed and LifeAct-RFP faithfully mark podocytes. Notably, we were able to demonstrate Ca2+ uptake or release into the podocyte cytoplasm, using the GCaMP6s fluorescent reporter, following high power laser illumination. It is possible that the laser light causes podocyte injury, both apoptosis and microtears, by mechanisms similar to those described previously in Drosophila epithelial wounds [39].
As nephrogenesis is a continuous process within the mesonephric adult kidney of zebrafish, it would not be unusual for podocyte cells at a range of developmental stages to be present. The paucity of early markers in the scRNA-seq data suggests that our selection protocol preferentially isolated more mature podocytes. This is to be expected since the reporters were driven by the podocin promoter.
By identifying established gene expression for young and mature podocytes, it was possible to attribute maturity and function to the majority of podocytes within clusters. We surmised that cluster 0 contained a subset of early podocytes, while clusters 0, 1 and 2 comprised maturing and developing podocytes. Clusters 3 and 4 contained podocytes which were likely to be earmarked for apoptosis. Pseudotime analysis largely confirmed our predictions.
Comparison of the zebrafish scRNA-seq data with transcription profiles of human and mouse podocyte libraries has revealed significant overlap in the respective transcription of genes associated with transcription factors, calcium ion binding proteins and actin filament assembly. This adds credence to the belief that the zebrafish is a suitable model for the study of renal function in healthy and diseased states. For example, the importance of lmx1bb, not only in podocyte progenitors but also in mature podocytes is borne out by observations in the Lmx1b knockout mouse. Knockout was lethal at birth, but inducible knockout in adults suggested that loss of Lmx1b leads to dysregulation of the actin cytoskeleton [40].
There were a number of compelling gene switches between clusters 0, 1, and 2, and clusters 3 and 4. These include the calcium ion-binding proteins, anxa13 and anxa2a (Fig. 4c), and the actin-binding proteins pfn2 and pfn1 (Fig. 4e). The apparent switch in expression of thymosin gene transcription between tmsb4x and tmsb1 (Fig. 4d) is interesting. Thymosin maintains the podocyte cytoskeleton. It has been shown in the mouse that loss of Tmsb4x worsens glomerular disease by increasing podocyte migration from the glomerular tuft to Bowman’s capsule [41]. It is possible that the switch in thymosin enhances the normal progression of podocytes as they age and are destined for removal. The same applies to the switch from timp2a to timp2b gene expression (Fig. 4f), which may lead to alterations in extracellular matrix deposition, breakdown and turnover with podocyte senescence [42].
A limitation of scRNA-seq is that it only shows which genes were active within the cells at the time of isolation. The gene expression data derived from this scRNA-seq experiment, however, provides a baseline and allows us to interrogate gene expression in the undamaged podocyte.
Conclusion
These data serve as a platform for future studies to compare normal zebrafish podocyte transcription profile with that of injured podocytes, for example following exposure to high power laser or puromycin amino-nucleoside (PAN) [16], which causes effacement and oedema, with a view to identifying indicators of injury, novel drug targets for repair and potentially the inhibition of biological pathways to prevent or slow injury progression.
We acknowledge Dr Charlotte Buckley, Mr Finn Bruton and Mr Aryan Kaveh for assistance with SPIM; Dr Cass Li and Dr John Wilson-Kanamori for assistance with bioinformatics analysis, and Dr Alessandro Brombin for assistance with construction of the zebrafish reference genome. For the movie shown in Supplementary Movie 1 we thank Eugene Katrukha (Utrecht University) and Lachlan Whitehead (WEHI) for their zoom macro. We also acknowledge Dr Carl Tucker and staff at the zebrafish facility. Fig. 1 was created using Mind the Graph platform and BioRender.com.
Author Contributions
Author contributions were as follows:
Conception or design of the work – SR, CB, LM, JM;
Acquisition or analysis or interpretation of data for the work – CB, LM, GM, KW, MB, SH;
Drafting work or revising it critically for important intellectual content – LM, CB, BC, JM.
Final approval of the version to be published – CB, LM, KW, GM, SH, MB, NH, BC, SR, JM.
Agreement to be accountable for all aspects of the work - CB, LM, KW, GM, SH, MB, NH, BC, SR, JM.
Funding Sources
CB and JM are supported by the British Heart Foundation Centre of Research Excellence Award (RE/08/001/23904); SH and KW by MRC/EPSRC DTA OPTIMA EP/L016559/1; LM by the Kidney Research UK (RP_026_20180305); GM is supported by the Medical Research Council, grant number MR/K015583/1 and Biotechnology and Biological Sciences Research Council, grant number BB/P02565X/1; NCH is supported by a Wellcome Trust Senior Research Fellowship in Clinical Science (ref. 219542/Z/19/Z).
Statement of Ethics
Animal experiments conform to internationally accepted ARRIVE standards and have been approved by the local institutional review body and the UK Home Office.
The authors have no conflicts of interest to declare.
| 1 Helmstadter M, Huber TB, Hermle T: Using the Drosophila Nephrocyte to Model Podocyte Function and Disease. Front Pediatr 2017;5:262. https://doi.org/10.3389/fped.2017.00262 |
||||
| 2 Wingert PTK, Jr., Rebecca A: Using Zebrafish to Study Podocyte Genesis During Kidney Development and Regeneration. Genesis 2015;27:320-331. | ||||
| 3 Ichimura K, Sakai T: Evolutionary morphology of podocytes and primary urine-producing apparatus. Anat Sci Int 2017;92:161-172. https://doi.org/10.1007/s12565-015-0317-7 |
||||
| 4 Reiser J, Altintas MM: Podocytes. F1000Res 2016;5:1-19. https://doi.org/10.12688/f1000research.7255.1 |
||||
| 5 Greka A, Mundel P: Cell biology and pathology of podocytes. Annu Rev Physiol 2012;74:299-323. https://doi.org/10.1146/annurev-physiol-020911-153238 |
||||
| 6 Kramer-Zucker AG, Wiessner S, Jensen AM, Drummond IA: Organization of the pronephric filtration apparatus in zebrafish requires Nephrin, Podocin and the FERM domain protein Mosaic eyes. Dev Biol 2005;285:316-329. https://doi.org/10.1016/j.ydbio.2005.06.038 |
||||
| 7 Schwarz K, Simons M, Reiser J, Saleem MA, Faul C, Kriz W, Shaw AS, Holzman LB, Mundel P: Podocin, a raft-associated component of the glomerular slit diaphragm, interacts with CD2AP and nephrin. J Clin Invest 2001;108:1621-1629. https://doi.org/10.1172/JCI200112849 |
||||
| 8 Martin CE, Jones N: Nephrin signaling in the podocyte: An updated view of signal regulation at the slit diaphragm and beyond. Front Endocrinol (Lausanne) 2018;9:1-12. https://doi.org/10.3389/fendo.2018.00302 |
||||
| 9 Merscher S, Pedigo CE, Mendez AJ: Metabolism, energetics, and lipid biology in the podocyte - Cellular cholesterol mediated glomerular injury. Front Endocrinol (Lausanne) 2014;5:1-11. https://doi.org/10.3389/fendo.2014.00169 |
||||
| 10 Giardino L, Armelloni S, Corbelli A, Mattinzoli D, Zennaro C, Guerrot D, Tourrel F, Ikehata M, Li M, Berra S, Carraro M, Messa P, Rastaldi MP: Podocyte glutamatergic signaling contributes to the function of the glomerular filtration barrier. J Am Soc Nephrol 2009;20:1929-1940. https://doi.org/10.1681/ASN.2008121286 |
||||
| 11 Wieder N, Greka A: Calcium, TRPC channels, and regulation of the actin cytoskeleton in podocytes: towards a future of targeted therapies. Pediatr Nephrol 2016;31:1047-1054. https://doi.org/10.1007/s00467-015-3224-1 |
||||
| 12 D'Agati VD: Pathobiology of focal segmental glomerulosclerosis. Curr Opin Nephrol Hypertens 2012;21:243-250. https://doi.org/10.1097/MNH.0b013e32835200df |
||||
| 13 Shankland SJ: The podocyte's response to injury: Role in proteinuria and glomerulosclerosis. Kidney Int 2006;69:2131-2147. https://doi.org/10.1038/sj.ki.5000410 |
||||
| 14 Ilatovskaya DV, Levchenko V, Lowing A, Shuyskiy LS, Palygin O, Staruschenko A: Podocyte injury in diabetic nephropathy: Implications of angiotensin II - dependent activation of TRPC channels. Sci Rep 2015;5:1-10. https://doi.org/10.1038/srep17637 |
||||
| 15 Nagata M: Podocyte injury and its consequences. Kidney Int 2016;89:1221-1230. https://doi.org/10.1016/j.kint.2016.01.012 |
||||
| 16 Rider SA, Bruton FA, Collins RG, Conway BR, Mullins JJ: The Efficacy of Puromycin and Adriamycin for Induction of Glomerular Failure in Larval Zebrafish Validated by an Assay of Glomerular Permeability Dynamics. Zebrafish 2018;15:234-242. https://doi.org/10.1089/zeb.2017.1527 |
||||
| 17 Westhoff JH, Steenbergen PJ, Thomas LSV, Heigwer J, Bruckner T, Cooper L, Tonshoff B, Hoffmann GF, Gehrig J: In vivo High-Content Screening in Zebrafish for Developmental Nephrotoxicity of Approved Drugs. Front Cell Dev Biol 2020;8:583. https://doi.org/10.3389/fcell.2020.00583 |
||||
| 18 Huang J, McKee M, Huang HD, Xiang A, Davidson AJ, Lu HA: A zebrafish model of conditional targeted podocyte ablation and regeneration. Kidney Int 2013;83:1193-1200. https://doi.org/10.1038/ki.2013.6 |
||||
| 19 Hansen KUI, Siegerist F, Daniel S, Schindler M, Iervolino A, Blumenthal A, Daniel C, Amann K, Zhou W, Endlich K, Endlich N: Prolonged podocyte depletion in larval zebrafish resembles mammalian focal and segmental glomerulosclerosis. FASEB J 2020;34:15961-15974. https://doi.org/10.1096/fj.202000724R |
||||
| 20 Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS: Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013;499:295-300. https://doi.org/10.1038/nature12354 |
||||
| 21 Badura A, Sun XR, Giovannucci A, Lynch LA, Wang SSH: Fast calcium sensor proteins for monitoring neural activity. Neurophotonics 2014;1:025008. https://doi.org/10.1117/1.NPh.1.2.025008 |
||||
| 22 Burford JL, Villanueva K, Lam L, Riquier-brison A, Hackl MJ, Pippin J, Shankland SJ, Peti-peterdi J: Intravital imaging of podocyte calcium in glomerular injury and disease. J Clin Invest 2014;124:2050-2058. https://doi.org/10.1172/JCI71702 |
||||
| 23 Belyy A, Merino F, Sitsel O, Raunser S: Structure of the Lifeact-F-actin complex. PLoS Biol 2020;18:e3000925. https://doi.org/10.1371/journal.pbio.3000925 |
||||
| 24 Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Bista M, Bradke F, Jenne D, Holak Ta, Werb Z, Sixt M, Wedlich-soldner R, Klopferspitz A, Avenue P, Francisco S: Lifeact: a versatile marker to visualize F-actin. Nat Methods 2010;5:1-8. https://doi.org/10.1038/nmeth.1220 |
||||
| 25 Westerfield M: A Guide for the Laboratory Use of Zebrafish Danio (Brachydanio) rerio, ed 4. Eugene, OR, University of Oregon Press, 2007. | ||||
| 26 He B, Ebarasi L, Hultenby K, Tryggvason K, Betsholtz C: Podocin-Green Fluorescence Protein Allows Visualization and Functional Analysis of Podocytes. J Am Soc Nephrol 2011;22:1019-1023. https://doi.org/10.1681/ASN.2010121291 |
||||
| 27 McConnell G, Trgrdh J, Amor R, Dempster J, Reid E, Amos WB: A novel optical microscope for imaging large embryos and tissue volumes with sub-cellular resolution throughout. eLife 2016;5:1-15. https://doi.org/10.7554/eLife.18659 |
||||
| 28 McConnell G, Amos WB: Application of the Mesolens for subcellular resolution imaging of intact larval and whole adult Drosophila. J Microsc 2018;270:252-258. https://doi.org/10.1111/jmi.12693 |
||||
| 29 Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A: Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676-682. https://doi.org/10.1038/nmeth.2019 |
||||
| 30 Buckley C, Carvalho MT, Young LK, Rider SA, McFadden C, Berlage C, Verdon RF, Taylor JM, Girkin JM, Mullins JJ: Precise spatio-temporal control of rapid optogenetic cell ablation with mem-KillerRed in Zebrafish. Sci Rep 2017;7:5096. https://doi.org/10.1038/s41598-017-05028-2 |
||||
| 31 Kaveh A, Bruton FA, Buckley C, Oremek MEM, Tucker CS, Mullins JJ, Taylor JM, Rossi AG, Denvir MA: Live Imaging of Heart Injury in Larval Zebrafish Reveals a Multi-Stage Model of Neutrophil and Macrophage Migration. Front Cell Dev Biol Biology 2020;8:1-22. https://doi.org/10.3389/fcell.2020.579943 |
||||
| 32 Butler A, Hoffman P, Smibert P, Papalexi E, Satija R: Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411-420. https://doi.org/10.1038/nbt.4096 |
||||
| 33 Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL: The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381-386. https://doi.org/10.1038/nbt.2859 |
||||
| 34 Tran T, Lindstrm NO, Ransick A, Brandine GDS, Kim AD, Der B, Peti-peterdi J, Smith AD, Grubbs B, McMahon JA, McMahon AP: In vivo developmental trajectories of human podocyte development inform in vitro differentiation of pluripotent stem- cell derived podocytes. Dev Cell 2019;50:102-116. https://doi.org/10.1016/j.devcel.2019.06.001 |
||||
| 35 Brunskill EW, Georgas K, Rumballe B, Little MH, Potter SS: Defining the molecular character of the developing and adult kidney podocyte. PLoS One 2011;6:1-12. https://doi.org/10.1371/journal.pone.0024640 |
||||
| 36 Kann M, Ettou S, Jung YL, Lenz MO, Taglienti ME, Park PJ, Schermer B, Benzing T, Kreidberg JA: Genome-wide analysis of Wilms' tumor 1-controlled gene expression in podocytes reveals key regulatory mechanisms. J Am Soc Nephrol 2015;26:2097-2104. https://doi.org/10.1681/ASN.2014090940 |
||||
| 37 Orrenius S, Zhivotovsky B, Nicotera P: Regulation of cell death: The calcium-apoptosis link. Nat Rev Mol Cell Biol 2003;4:552-565. https://doi.org/10.1038/nrm1150 |
||||
| 38 Orrenius S, Gogvadze V, Zhivotovsky B: Calcium and mitochondria in the regulation of cell death. Biochem Biophys Res Commun 2015;460:72-81. https://doi.org/10.1016/j.bbrc.2015.01.137 |
||||
| 39 Shannon EK, Stevens A, Edrington W, Zhao Y, Jayasinghe AK, Page-McCaw A, Hutson MS: Multiple Mechanisms Drive Calcium Signal Dynamics around Laser-Induced Epithelial Wounds. Biophys J 2017;113:1623-1635. https://doi.org/10.1016/j.bpj.2017.07.022 |
||||
| 40 Burghardt T, Kastner J, Suleiman H, Rivera-Milla E, Stepanova N, Lottaz C, Kubitza M, Bger CA, Schmidt S, Gorski Ma: LMX1B is essential for the maintenance of differentiated podocytes in adult kidneys. J Am Soc Nephrol 2013;24:1830-1848. https://doi.org/10.1681/ASN.2012080788 |
||||
| 41 Vasilopoulou E, Kolatsi-Joannou M, Lindenmeyer MT, White KE, Robson MG, Cohen CD, Sebire NJ, Riley PR, Winyard PJ, Long DA: Loss of endogenous thymosin beta4 accelerates glomerular disease. Kidney Int 2016;90:1056-1070. https://doi.org/10.1016/j.kint.2016.06.032 |
||||
| 42 Thrailkill KM, Bunn RC, Fowlkes JL: Matrix metalloproteinases: their potential role in the pathogenesis of diabetic nephropathy. Endocrine 2009;35:1-7. https://doi.org/10.1007/s12020-008-9114-6 |
||||