Corresponding Author: Meltem Weger and Frédéric Gachon
Institute for Molecular Bioscience, The University of Queensland, St. Lucia, QLD, 4072 (Australia)
E-Mail m.weger@uq.edu.au; f.gachon@uq.edu.au
The Mechanisms and Physiological Consequences of Diurnal Hepatic Cell Size Fluctuations: A Brief Review
Meltem Wegera Benjamin D. Wegera Frédéric Gachona
aInstitute for Molecular Bioscience, The University of Queensland, St. Lucia, QLD, Australia
Introduction
The circadian clock is an evolutionarily conserved, endogenous mechanism which allows organisms to anticipate the daily changes in their environment due to the light/dark cycles resulting from the Earth’s rotation around its own axis. The circadian clock regulates many aspects of an organisms’ behaviour such as feeding/fasting cycles and physiology. Mammalian circadian clocks are hierarchically organised with a central clock located in the hypothalamic suprachiasmatic nuclei (SCN) [1, 2] and peripheral clocks present in peripheral tissues such as the liver [3-6]. Similar to the circadian clock of SCN neurons, peripheral clocks are self-sustained and autonomous. However, to adjust to daily environmental changes and to ensure phase coherence between body clocks, peripheral clocks are synchronized by the SCN master clock [3] which is in turn synchronized by environmental light received by the retina [7, 8]. On a molecular level, circadian clocks consist of interconnected transcriptional and translational feedback loops that regulate rhythmic gene expression with a 24 h cycle. The core loop consists of the BMAL1 (or ARNTL) transcription factor that, once heterodimerized with CLOCK or NPAS2, binds to E-box like elements in the promoters of circadian clock target genes to induce their expression [9, 10]. This includes also Period (Per) and Cryptochromes (Cry) [9, 11], which after their translation and accumulation in the cytosol, translocate into the nucleus and inhibit BMAL1 transcription, resulting in the inhibition of BMAL1-driven circadian clock target genes [12-14]. For a more in-depth review of the circadian clock, see [15, 16].
Of particular note, the circadian clock plays a key role in regulating metabolism. Many metabolic processes that take place in the liver, including glucose, bile acid and lipid metabolism, as well as hepatic detoxification, are coordinated with feeding/fasting cycles and are subject to circadian clock regulation [17-19]. Furthermore, the liver is also a secretory organ that synthesizes and secretes most blood proteins such as albumin, coagulation factors, and complement proteins involved in the immune response. Interestingly, most of these proteins exhibit rhythmic levels in the blood of both rodents [20, 21] and human [22, 23]. However, the role of the circadian clock on the secretion of these proteins seems limited, with rather a critical role of feeding rhythms on the regulation of this secretion [24].
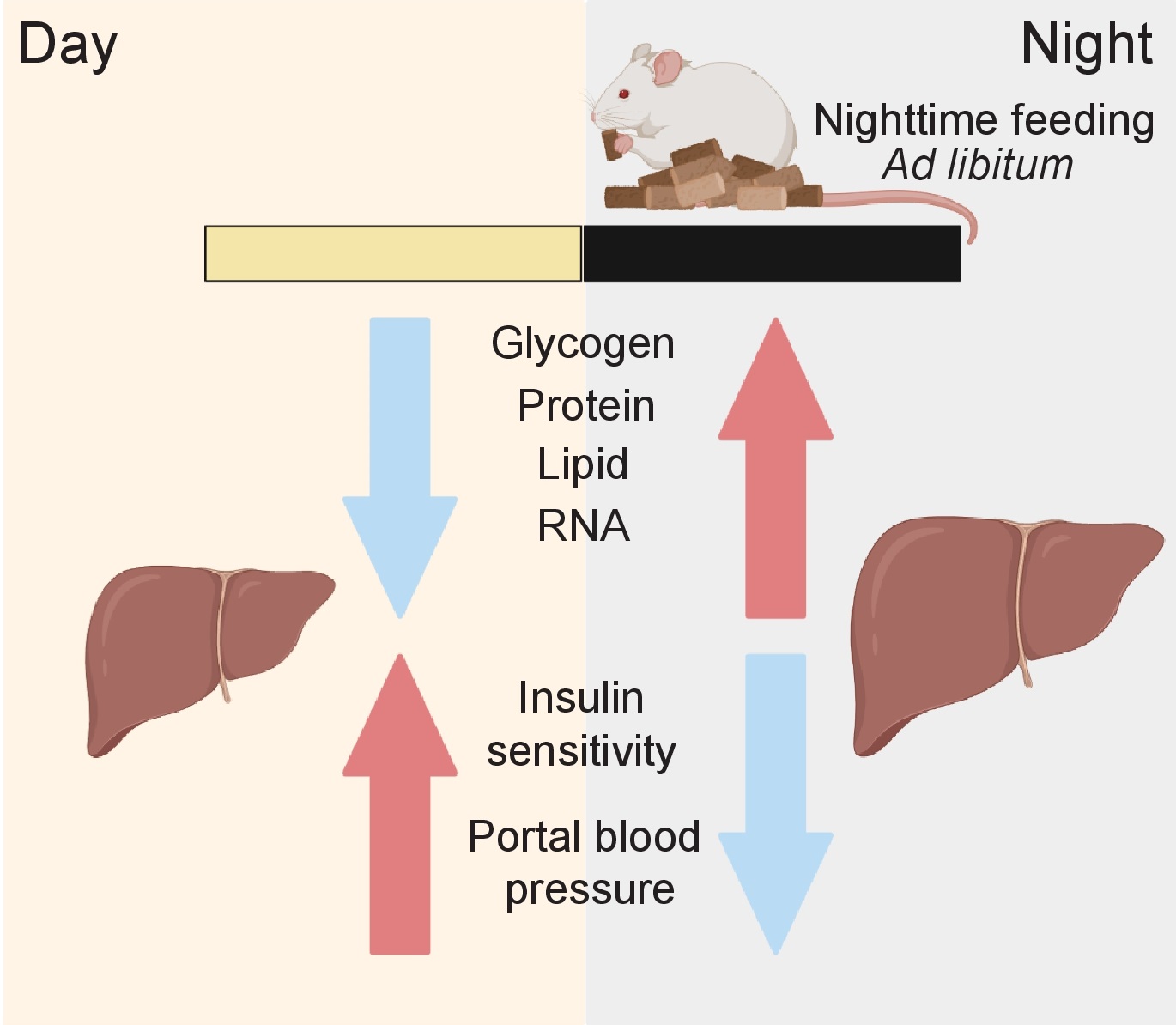
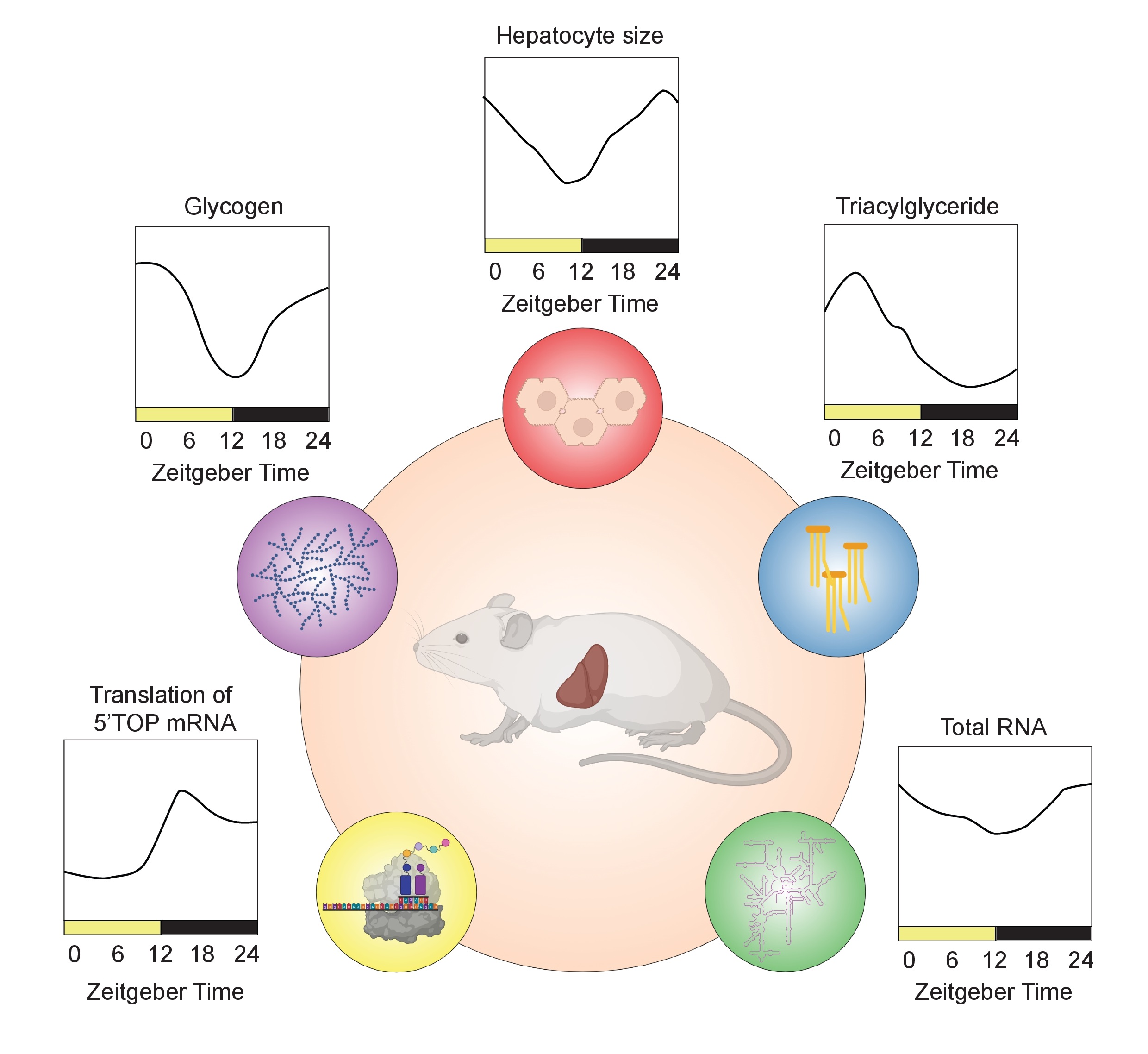
The genes and proteins involved in these processes are orchestrated by environmental cues via the SCN master clock and/or by the cell-autonomous peripheral liver clock itself [15, 25, 26]. Circadian regulation of liver function occurs on multiple regulatory levels of gene expression, including transcription [27-29], post-transcriptional regulation [30, 31], mRNA translation [32-34], post-translational regulation [35, 36], protein trafficking [37], and secretion [24]. Increasing evidence associates liver gene and protein expression with daily (or diurnal) fluctuations in liver size (Fig. 1). The exact role of this phenomenon is unknown but might be related to physiological organ functions. It thus might be of medical interest as it can serve as a readout for a normal and “healthy” organ status. For example, hepatocyte size fluctuations also correlate with liver portal blood pressure suggesting an impact of the volume of the hepatocyte compartment on the size of blood vessels [38, 39] (Fig. 1 and 2). Moreover, an increased liver/hepatocyte size and lower size fluctuations have been also associated with cirrhosis and chronic alcohol consumption [40, 41]. However, the factors and mechanisms contributing to liver size fluctuations remain to date poorly understood. That said, recent evidence suggests that daily fluctuations in liver size are regulated by the circadian clock and the associated feeding/fasting cycle. Here, we provide an overview of the current literature supporting this idea and summarize the body of work dealing with the potential underlying molecular mechanisms, including the role of hepatic macromolecule content and protein synthesis and changes in hepatocyte cell volume via osmotic regulation.
Potential mechanisms for feeding dependent hepatocyte fluctuations
Though it is clear that the feeding/fasting cycle plays an important role in hepatocyte and liver size fluctuations, the underlying mechanisms have yet to be determined. One possibility discussed below is that hepatocyte and liver size changes may be a result of daily fluctuations in hepatic macromolecule content, but it is still unclear whether this goes beyond correlation.
The role of macromolecules and protein synthesis
In synchrony with liver size, lipid content (an integral part of the cell plasma membrane), glycogen (the storage form of glucose), ribosomal RNA, and proteins also fluctuate throughout the day [37, 43, 47] (Fig. 1 and 2). As proteins constitute most of the liver’s dry mass, and protein synthesis that peak at night [34, 47], cellular fluctuation in protein content may mainly contribute to changes in liver size [47]. The number of ribosomes has been observed to follow a diurnal pattern similar to protein content [34, 47, 62], suggesting that the ribosome number is critical for translation efficiency and is rate limiting for hepatic protein synthesis [34, 47]. In fact, both ribosomal proteins and ribosomal RNAs show diurnal fluctuations in the liver through a mechanism involving their rhythmic synthesis [34, 47] and polyadenylation-dependent degradation [47]. The decrease in protein content during the day might be also a result of rhythmic protein degradation via autophagy, likely to compensate for the decrease in nutrient intake during the sleep/fasting period [63]. The recently described fasting-induced specific degradation of ribosomes via ribophagy might also play a role in the decreased protein content during the day [64]. Thus, ribosome synthesis/degradation might be a way to store an excess of food-derived amino acids and nucleotides during the night (murine activity phase) and use them during the day (murine resting phase) to maintain constant levels, in a similar fashion as glycogen synthesis and breakdown keeps blood glucose at constant level. In this context, fluctuations in hepatocyte size would be a concomitant feature that is driven by daily changes in hepatic protein synthesis and content. This would be in line with the concept that protein synthesis, a highly energy-dependent process, is actively restricted to times when enough energy and nutrients are available.
The role of osmotic cell regulation of hepatic cell volume
It is well-established that virtually all vertebrate cells have the capacity to actively undergo changes in cell volume through cell swelling (also called hydration) and shrinkage which is based on their ability to take up or release cellular ions and organic osmolytes. The dynamics of these processes have been described to occur on a time scale of few minutes. However, it is tempting to speculate that at least some of the suggested factors implicated in osmotic cell regulation and their linked functions [65, 66] might play a role in the diurnal regulation of cell and liver size. For example, cytoskeletal elements involved in the organization of the microtubular structure or the actin cytoskeleton, characterized by the ratio between F and G actin, are remodelled by cell volume changes [67, 68] and contribute to cell swelling-dependent proteolysis [69, 70]. Accordingly, they also exhibit diurnal dynamics in mouse liver [45], suggesting that they may be involved in this process. In fact, there is some evidence that daily fluctuations in hepatocyte cell volume may actually drive changes in macromolecule content rather than the other way around. Although the precise molecular mechanisms are still poorly understood, changes in cell volume are known to play a crucial role in cell physiology, modulating both gene expression and metabolism [65, 66] including the synthesis and degradation of glycogens [71-73] and proteins [74-76]. More research is needed to answer the question of how changes in cell volume and macromolecule content is related to diurnal regulation of hepatocyte size and liver size.
Insulin/glucagon and mTOR signalling as potential mediators
Insulin/glucagon signalling is an important feeding-dependent regulator of metabolism [77], and previous research suggests that it is important for liver size fluctuations. The liver-specific loss of the insulin receptor is known to impact liver size [78, 79], and the rhythmic secretion of insulin and glucagon is subject to circadian clock regulation [80-82]. Moreover, this pathway has been identified as a crucial regulator of hepatic cell volume and linked functions. For example, insulin stimulates the cellular retention of K+ and thus allows hepatocyte swelling though an influx of water which mediates the inhibition of proteolysis and glycogen synthesis [83, 84], while glucagon counters the effects of insulin [84, 85]. In turn, fluctuations in liver size also influences the liver’s insulin response, as insulin sensitivity is time-of-day and circadian clock-dependent, and maximum insulin actions occur when the liver has reached its smallest size [86, 87]. Interestingly, the inactivation of the 3-phosphoinositide-dependent protein kinase-1 (PDK1) that is involved in the regulation of ion transport during hepatocyte shrinkage [88, 89] also results in an increase in insulin sensitivity [90]. Conversely, inactivation of the serum- and glucocorticoid-regulated protein kinase 1 (SGK1) is involved in cell swelling [66] and is associated with insulin resistance [91]. Together, these results suggest that insulin plays an important role in the regulation of liver cell size which in turn impacts insulin sensitivity.
In addition to insulin/glucagon signalling, the mTOR (mechanistic Target of Rapamycin) signalling pathway has also been implicated as a regulator of hepatic cell volume and linked functions [92, 93]. It thus might be another important driver of daily liver size fluctuations. Activation of the mTOR signalling pathway, through the activity of complex 1 (mTORC1) and 2 (mTORC2), is a well-known nutrient responsive regulator that plays a crucial role in eukaryotic cell growth, metabolism, and mRNA translation/protein synthesis [94]. In line with mTOR’s role in these processes, an inhibition or loss of mTOR factors reduces liver size in mice, whereas its activation leads to an increased liver size [79, 95, 96]. Notably, the activation of the mTOR signalling pathway is subject to circadian clock regulation [34, 35, 97, 98] and is involved in the rhythmic regulation of the translation of ribosomal proteins and translation regulation factors [32]. In this way, the mTOR signalling pathway drives rhythmic ribosome biogenesis and protein synthesis [34]. Nevertheless, the interconnection between the circadian clock and the mTOR pathway is poorly understood and further research is needed. Previous work has shown that BMAL1 is phosphorylated by the mTOR-effector kinase S6K1, leading to the regulation of mRNA translation by directly interacting with the translational machinery [99]. In line with this study, the negative regulator of BMAL1 activity, PER2, reportedly modulates mTORC1 activity in response to fasting in cell culture studies and thus could regulate BMAL1-induced protein translation [100]. Although the idea that the circadian clock can directly impact protein translation via the core clock members BMAL1 [99] and PER2 [100] is appealing, some discrepancies need to be considered. First, most genes (approximately 70%) that are rhythmically translated in mouse liver rely on rhythmic mRNA levels. The rhythmic translation of these genes is a result of a rhythmic transcription rather than an active rhythmic regulation of translation [32, 34]. Second, investigations in Bmal1 knockout mice fail to confirm a predominant role of BMAL1 in rhythmic translation in the liver. Indeed, relative translation efficiency is globally conserved, and only a minor fraction of mRNAs (with the exception of mRNA encoding for ribosomal proteins) are differentially translated in the liver of Bmal1 knockout mice compared with their wild-type littermates [32]. Thus, the modulation of the mTOR pathway by the circadian clock may instead occur indirectly through the regulation of the feeding/fasting cycle [32, 51], rRNA transcription and stability [34, 47], and protein transport into the nucleus [37].
Conclusion
As discussed here, liver size fluctuations correlate with daily changes in nutrient uptake, storage, and metabolism. Liver size increases with nutrient intake and storage during the active/feeding period and decreases when stored nutrients are broken down during the sleep/fasting period (Fig. 1 and 2). However, these size fluctuations appear to be a more complex process rather than simply a passive “dilatation” of the liver in response to macromolecule synthesis and storage; additional mechanisms of cell volume regulation are likely involved as well. These mechanisms may potentially even be actively driving liver size fluctuations. However, many questions remain and further research will be required to answer these complex questions and to obtain a better understanding of the underlying mechanisms.
Although this review article has focused on the liver, it is possible that the mechanisms discussed here might also be relevant for other organs: daily fluctuations in size or volume have also been reported in the brain [101-103], eye [104, 105], spleen [106], pancreatic β-cells [107], and epithelial cells of the intestine [108]. With the improvement of in vivo imaging techniques, it would be interesting to measure daily changes in tissues’ size in both healthy and pathological conditions, or in conditions of perturbed circadian rhythms as during shift work. This would help to understand their physiological functions and define potential new biological markers of multiple disease states.
B.D.W is supported by an UQ Early Career Researcher Grant (UQECR2058233) and F.G. receives support from the Institute for Molecular Bioscience, The University of Queensland.
The authors declare they have no conflict of interests.
| 1 Stephan FK, Zucker I: Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A 1972;69:1583-1586. https://doi.org/10.1073/pnas.69.6.1583 |
||||
| 2 Moore RY, Eichler VB: Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res 1972;42:201-206. https://doi.org/10.1016/0006-8993(72)90054-6 |
||||
| 3 Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS: PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 2004;101:5339-5346. https://doi.org/10.1073/pnas.0308709101 |
||||
| 4 Balsalobre A, Damiola F, Schibler U: A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 1998;93:929-937. https://doi.org/10.1016/S0092-8674(00)81199-X |
||||
| 5 Tosini G, Menaker M: Circadian rhythms in cultured mammalian retina. Science 1996;272:419-421. https://doi.org/10.1126/science.272.5260.419 |
||||
| 6 Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H: Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000;288:682-685. https://doi.org/10.1126/science.288.5466.682 |
||||
| 7 Meijer JH, Thio B, Albus H, Schaap J, Ruijs AC: Functional absence of extraocular photoreception in hamster circadian rhythm entrainment. Brain Res 1999;831:337-339. https://doi.org/10.1016/S0006-8993(99)01509-7 |
||||
| 8 Hull JT, Czeisler CA, Lockley SW: Suppression of Melatonin Secretion in Totally Visually Blind People by Ocular Exposure to White Light: Clinical Characteristics. Ophthalmology 2018;125:1160-1171. https://doi.org/10.1016/j.ophtha.2018.01.036 |
||||
| 9 Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ: Role of the CLOCK protein in the mammalian circadian mechanism. Science 1998;280:1564-1569. https://doi.org/10.1126/science.280.5369.1564 |
||||
| 10 Yoo SH, Ko CH, Lowrey PL, Buhr ED, Song EJ, Chang S, Yoo OJ, Yamazaki S, Lee C, Takahashi JS: A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo . Proc Natl Acad Sci U S A 2005;102:2608-2613. https://doi.org/10.1073/pnas.0409763102 |
||||
| 11 Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM: mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 1999;98:193-205. https://doi.org/10.1016/S0092-8674(00)81014-4 |
||||
| 12 Griffin EA, Jr., Staknis D, Weitz CJ: Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 1999;286:768-771. https://doi.org/10.1126/science.286.5440.768 |
||||
| 13 Sangoram AM, Saez L, Antoch MP, Gekakis N, Staknis D, Whiteley A, Fruechte EM, Vitaterna MH, Shimomura K, King DP, Young MW, Weitz CJ, Takahashi JS: Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron 1998;21:1101-1113. https://doi.org/10.1016/S0896-6273(00)80627-3 |
||||
| 14 Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB: Feedback repression is required for mammalian circadian clock function. Nat Genet 2006;38:312-319. https://doi.org/10.1038/ng1745 |
||||
| 15 Schibler U, Gotic I, Saini C, Gos P, Curie T, Emmenegger Y, Sinturel F, Gosselin P, Gerber A, Fleury-Olela F, Rando G, Demarque M, Franken P: Clock-Talk: Interactions between Central and Peripheral Circadian Oscillators in Mammals. Cold Spring Harb Symp Quant Biol 2015;80:223-232. https://doi.org/10.1101/sqb.2015.80.027490 |
||||
| 16 Rijo-Ferreira F, Takahashi JS: Genomics of circadian rhythms in health and disease. Genome Med 2019;11:82. https://doi.org/10.1186/s13073-019-0704-0 |
||||
| 17 Atger F, Mauvoisin D, Weger B, Gobet C, Gachon F: Regulation of Mammalian Physiology by Interconnected Circadian and Feeding Rhythms. Front Endocrinol (Lausanne) 2017;8:42. https://doi.org/10.3389/fendo.2017.00042 |
||||
| 18 Reinke H, Asher G: Crosstalk between metabolism and circadian clocks. Nat Rev Mol Cell Biol 2019;20:227-241. https://doi.org/10.1038/s41580-018-0096-9 |
||||
| 19 Panda S: Circadian physiology of metabolism. Science 2016;354:1008-1015. https://doi.org/10.1126/science.aah4967 |
||||
| 20 Scheving LE, Pauly JE, Tsai TH: Circadian fluctuation in plasma proteins of the rat. Am J Physiol 1968;215:1096-1101. https://doi.org/10.1152/ajplegacy.1968.215.5.1096 |
||||
| 21 Scheving LE, Pauly JE: Daily rhythmic variations in blood coagulation times in rats. Anat Rec 1967;157:657-665. https://doi.org/10.1002/ar.1091570411 |
||||
| 22 Kapiotis S, Jilma B, Quehenberger P, Ruzicka K, Handler S, Speiser W: Morning hypercoagulability and hypofibrinolysis. Diurnal variations in circulating activated factor VII, prothrombin fragment F1+2, and plasmin-plasmin inhibitor complex. Circulation 1997;96:19-21. https://doi.org/10.1161/01.CIR.96.1.19 |
||||
| 23 Haus E, Cusulos M, Sackett-Lundeen L, Swoyer J: Circadian variations in blood coagulation parameters, alpha-antitrypsin antigen and platelet aggregation and retention in clinically healthy subjects. Chronobiol Int 1990;7:203-216. https://doi.org/10.3109/07420529009056976 |
||||
| 24 Mauvoisin D, Wang J, Jouffe C, Martin E, Atger F, Waridel P, Quadroni M, Gachon F, Naef F: Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc Natl Acad Sci U S A 2014;111:167-172. https://doi.org/10.1073/pnas.1314066111 |
||||
| 25 Johnson BP, Walisser JA, Liu Y, Shen AL, McDearmon EL, Moran SM, McIntosh BE, Vollrath AL, Schook AC, Takahashi JS, Bradfield CA: Hepatocyte circadian clock controls acetaminophen bioactivation through NADPH-cytochrome P450 oxidoreductase. Proc Natl Acad Sci U S A 2014;111:18757-18762. https://doi.org/10.1073/pnas.1421708111 |
||||
| 26 Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U: System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 2007;5:e34. https://doi.org/10.1371/journal.pbio.0050034 |
||||
| 27 Yeung J, Mermet J, Jouffe C, Marquis J, Charpagne A, Gachon F, Naef F: Transcription factor activity rhythms and tissue-specific chromatin interactions explain circadian gene expression across organs. Genome Res 2018;28:182-191. https://doi.org/10.1101/gr.222430.117 |
||||
| 28 Menet JS, Rodriguez J, Abruzzi KC, Rosbash M: Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife 2012;1:e00011. https://doi.org/10.7554/eLife.00011 |
||||
| 29 Weger BD, Gobet C, David FPA, Atger F, Martin E, Phillips NE, Charpagne A, Weger M, Naef F, Gachon F: Systematic analysis of differential rhythmic liver gene expression mediated by the circadian clock and feeding rhythms. Proc Natl Acad Sci U S A 2021;118:e2015803118. https://doi.org/10.1073/pnas.2015803118 |
||||
| 30 Gotic I, Omidi S, Fleury-Olela F, Molina N, Naef F, Schibler U: Temperature regulates splicing efficiency of the cold-inducible RNA-binding protein gene Cirbp. Genes Dev 2016;30:2005-2017. https://doi.org/10.1101/gad.287094.116 |
||||
| 31 Kojima S, Sher-Chen EL, Green CB: Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev 2012;26:2724-2736. https://doi.org/10.1101/gad.208306.112 |
||||
| 32 Atger F, Gobet C, Marquis J, Martin E, Wang J, Weger B, Lefebvre G, Descombes P, Naef F, Gachon F: Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci U S A 2015;112:E6579-6588. https://doi.org/10.1073/pnas.1515308112 |
||||
| 33 Janich P, Arpat AB, Castelo-Szekely V, Lopes M, Gatfield D: Ribosome profiling reveals the rhythmic liver translatome and circadian clock regulation by upstream open reading frames. Genome Res 2015;25:1848-1859. https://doi.org/10.1101/gr.195404.115 |
||||
| 34 Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F: The circadian clock coordinates ribosome biogenesis. PLoS Biol 2013;11:e1001455. https://doi.org/10.1371/journal.pbio.1001455 |
||||
| 35 Robles MS, Humphrey SJ, Mann M: Phosphorylation Is a Central Mechanism for Circadian Control of Metabolism and Physiology. Cell Metab 2017;25:118-127. https://doi.org/10.1016/j.cmet.2016.10.004 |
||||
| 36 Mauvoisin D, Gachon F: Proteomics in Circadian Biology. J Mol Biol 2019;432:3565-3577. https://doi.org/10.1016/j.jmb.2019.12.004 |
||||
| 37 Wang J, Mauvoisin D, Martin E, Atger F, Galindo AN, Dayon L, Sizzano F, Palini A, Kussmann M, Waridel P, Quadroni M, Dulic V, Naef F, Gachon F: Nuclear Proteomics Uncovers Diurnal Regulatory Landscapes in Mouse Liver. Cell Metab 2017;25:102-117. https://doi.org/10.1016/j.cmet.2016.10.003 |
||||
| 38 Garcia-Pagan JC, Feu F, Castells A, Luca A, Hermida RC, Rivera F, Bosch J, Rodes J: Circadian variations of portal pressure and variceal hemorrhage in patients with cirrhosis. Hepatology 1994;19:595-601. https://doi.org/10.1002/hep.1840190309 |
||||
| 39 Orrego H, Blendis LM, Crossley IR, Medline A, Macdonald A, Ritchie S, Israel Y: Correlation of intrahepatic pressure with collagen in the Disse space and hepatomegaly in humans and in the rat. Gastroenterology 1981;80:546-556. https://doi.org/10.1016/0016-5085(81)90018-4 |
||||
| 40 Blendis LM, Orrego H, Crossley IR, Blake JE, Medline A, Isreal Y: The role of hepatocyte enlargement in hepatic pressure in cirrhotic and noncirrhotic alcoholic liver disease. Hepatology 1982;2:539-546. https://doi.org/10.1002/hep.1840020505 |
||||
| 41 Leung NW, Farrant P, Peters TJ: Liver volume measurement by ultrasonography in normal subjects and alcoholic patients. J Hepatol 1986;2:157-164. https://doi.org/10.1016/S0168-8278(86)80074-5 |
||||
| 42 Fisher HI BL: Diurnal cycles in liver weights in birds. . Condor 1957;59:364-372. https://doi.org/10.2307/1365247 |
||||
| 43 Wilson WO, McFarland LZ: Diurnal changes in livers and digestive systems of coturnix as related to three photoperiodic regimens. Poult Sci 1969;48:477-482. https://doi.org/10.3382/ps.0480477 |
||||
| 44 Liu Y, Basty N, Whitcher B, Bell JD, Sorokin EP, van Bruggen N, Thomas EL, Cule M: Genetic architecture of 11 organ traits derived from abdominal MRI using deep learning. Elife 2021;10:e65554. https://doi.org/10.7554/eLife.65554 |
||||
| 45 Gerber A, Esnault C, Aubert G, Treisman R, Pralong F, Schibler U: Blood-borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell 2013;152:492-503. https://doi.org/10.1016/j.cell.2012.12.027 |
||||
| 46 Stanger BZ: Cellular homeostasis and repair in the mammalian liver. Annu Rev Physiol 2015;77:179-200. https://doi.org/10.1146/annurev-physiol-021113-170255 |
||||
| 47 Sinturel F, Gerber A, Mauvoisin D, Wang J, Gatfield D, Stubblefield JJ, Green CB, Gachon F, Schibler U: Diurnal Oscillations in Liver Mass and Cell Size Accompany Ribosome Assembly Cycles. Cell 2017;169:651-663 e614. https://doi.org/10.1016/j.cell.2017.04.015 |
||||
| 48 Challet E: The circadian regulation of food intake. Nat Rev Endocrinol 2019;15:393-405. https://doi.org/10.1038/s41574-019-0210-x |
||||
| 49 Agius L, Peak M, al-Habori M: What determines the increase in liver cell volume in the fasted-to-fed transition: glycogen or insulin? Biochem J 1991;276:843-845. https://doi.org/10.1042/bj2760843 |
||||
| 50 Diaz-Munoz M, Vazquez-Martinez O, Baez-Ruiz A, Martinez-Cabrera G, Soto-Abraham MV, Avila-Casado MC, Larriva-Sahd J: Daytime food restriction alters liver glycogen, triacylglycerols, and cell size. A histochemical, morphometric, and ultrastructural study. Comp Hepatol 2010;9:5. https://doi.org/10.1186/1476-5926-9-5 |
||||
| 51 Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, Panda S: Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 2012;15:848-860. https://doi.org/10.1016/j.cmet.2012.04.019 |
||||
| 52 Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U: Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 2000;14:2950-2961. https://doi.org/10.1101/gad.183500 |
||||
| 53 Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S: Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A 2009;106:21453-21458. https://doi.org/10.1073/pnas.0909591106 |
||||
| 54 Mange F, Praz V, Migliavacca E, Willis IM, Schutz F, Hernandez N, Cycli XC: Diurnal regulation of RNA polymerase III transcription is under the control of both the feeding-fasting response and the circadian clock. Genome Res 2017;27:973-984. https://doi.org/10.1101/gr.217521.116 |
||||
| 55 Greenwell BJ, Trott AJ, Beytebiere JR, Pao S, Bosley A, Beach E, Finegan P, Hernandez C, Menet JS: Rhythmic Food Intake Drives Rhythmic Gene Expression More Potently than the Hepatic Circadian Clock in Mice. Cell Rep 2019;27:649-657.e5. https://doi.org/10.1016/j.celrep.2019.03.064 |
||||
| 56 Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J: Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005;308:1043-1045. https://doi.org/10.1126/science.1108750 |
||||
| 57 Lamia KA, Storch KF, Weitz CJ: Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A 2008;105:15172-15177. https://doi.org/10.1073/pnas.0806717105 |
||||
| 58 Adamovich Y, Rousso-Noori L, Zwighaft Z, Neufeld-Cohen A, Golik M, Kraut-Cohen J, Wang M, Han X, Asher G: Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab 2014;19:319-330. https://doi.org/10.1016/j.cmet.2013.12.016 |
||||
| 59 van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A: Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 1999;398:627-630. https://doi.org/10.1038/19323 |
||||
| 60 Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA: Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 2000;103:1009-1017. https://doi.org/10.1016/S0092-8674(00)00205-1 |
||||
| 61 Nagai K, Nishio T, Nakagawa H, Nakamura S, Fukuda Y: Effect of bilateral lesions of the suprachiasmatic nuclei on the circadian rhythm of food-intake. Brain Res 1978;142:384-389. https://doi.org/10.1016/0006-8993(78)90648-0 |
||||
| 62 Fishman B, Wurtman RJ, Munro HN: Daily rhythms in hepatic polysome profiles and tyrosine transaminase activity: role of dietary protein. Proc Natl Acad Sci U S A 1969;64:677-682. https://doi.org/10.1073/pnas.64.2.677 |
||||
| 63 Ryzhikov M, Ehlers A, Steinberg D, Xie W, Oberlander E, Brown S, Gilmore PE, Townsend RR, Lane WS, Dolinay T, Nakahira K, Choi AMK, Haspel JA: Diurnal Rhythms Spatially and Temporally Organize Autophagy. Cell Rep 2019;26:1880-1892.e6. https://doi.org/10.1016/j.celrep.2019.01.072 |
||||
| 64 Wyant GA, Abu-Remaileh M, Frenkel EM, Laqtom NN, Dharamdasani V, Lewis CA, Chan SH, Heinze I, Ori A, Sabatini DM: NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 2018;360:751-758. https://doi.org/10.1126/science.aar2663 |
||||
| 65 Hoffmann EK, Lambert IH, Pedersen SF: Physiology of cell volume regulation in vertebrates. Physiol Rev 2009;89:193-277. https://doi.org/10.1152/physrev.00037.2007 |
||||
| 66 Lang PA, Graf D, Boini KM, Lang KS, Klingel K, Kandolf R, Lang F: Cell volume, the serum and glucocorticoid inducible kinase 1 and the liver. Z Gastroenterol 2011;49:713-719. https://doi.org/10.1055/s-0031-1273425 |
||||
| 67 Theodoropoulos PA, Stournaras C, Stoll B, Markogiannakis E, Lang F, Gravanis A, Haussinger D: Hepatocyte swelling leads to rapid decrease of the G-/total actin ratio and increases actin mRNA levels. FEBS Lett 1992;311:241-245. https://doi.org/10.1016/0014-5793(92)81111-X |
||||
| 68 Schulz WA, Eickelmann P, Hallbrucker C, Sies H, Haussinger D: Increase of beta-actin mRNA upon hypotonic perfusion of perfused rat liver. FEBS Lett 1991;292:264-266. https://doi.org/10.1016/0014-5793(91)80880-C |
||||
| 69 vom Dahl S, Stoll B, Gerok W, Haussinger D: Inhibition of proteolysis by cell swelling in the liver requires intact microtubular structures. Biochem J 1995;308:529-536. https://doi.org/10.1042/bj3080529 |
||||
| 70 Busch GL, Schreiber R, Dartsch PC, Volkl H, Vom Dahl S, Haussinger D, Lang F: Involvement of microtubules in the link between cell volume and pH of acidic cellular compartments in rat and human hepatocytes. Proc Natl Acad Sci U S A 1994;91:9165-9169. https://doi.org/10.1073/pnas.91.19.9165 |
||||
| 71 Peak M, al-Habori M, Agius L: Regulation of glycogen synthesis and glycolysis by insulin, pH and cell volume. Interactions between swelling and alkalinization in mediating the effects of insulin. Biochem J 1992;282:797-805. https://doi.org/10.1042/bj2820797 |
||||
| 72 al-Habori M, Peak M, Thomas TH, Agius L: The role of cell swelling in the stimulation of glycogen synthesis by insulin. Biochem J 1992;282:789-796. https://doi.org/10.1042/bj2820789 |
||||
| 73 Baquet A, Hue L, Meijer AJ, van Woerkom GM, Plomp PJ: Swelling of rat hepatocytes stimulates glycogen synthesis. J Biol Chem 1990;265:955-959. https://doi.org/10.1016/S0021-9258(19)40142-7 |
||||
| 74 Stoll B, Gerok W, Lang F, Haussinger D: Liver cell volume and protein synthesis. Biochem J 1992;287:217-222. https://doi.org/10.1042/bj2870217 |
||||
| 75 Haussinger D, Hallbrucker C, vom Dahl S, Decker S, Schweizer U, Lang F, Gerok W: Cell volume is a major determinant of proteolysis control in liver. FEBS Lett 1991;283:70-72. https://doi.org/10.1016/0014-5793(91)80556-I |
||||
| 76 Vom Dahl S, Haussinger D: Nutritional state and the swelling-induced inhibition of proteolysis in perfused rat liver. J Nutr 1996;126:395-402. https://doi.org/10.1093/jn/126.2.395 |
||||
| 77 Petersen MC, Vatner DF, Shulman GI: Regulation of hepatic glucose metabolism in health and disease. Nat Rev Endocrinol 2017;13:572-587. https://doi.org/10.1038/nrendo.2017.80 |
||||
| 78 Michael MD, Kulkarni RN, Postic C, Previs SF, Shulman GI, Magnuson MA, Kahn CR: Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell 2000;6:87-97. https://doi.org/10.1016/S1097-2765(05)00015-8 |
||||
| 79 Kucejova B, Duarte J, Satapati S, Fu X, Ilkayeva O, Newgard CB, Brugarolas J, Burgess SC: Hepatic mTORC1 Opposes Impaired Insulin Action to Control Mitochondrial Metabolism in Obesity. Cell Rep 2016;16:508-519. https://doi.org/10.1016/j.celrep.2016.06.006 |
||||
| 80 Pulimeno P, Mannic T, Sage D, Giovannoni L, Salmon P, Lemeille S, Giry-Laterriere M, Unser M, Bosco D, Bauer C, Morf J, Halban P, Philippe J, Dibner C: Autonomous and self-sustained circadian oscillators displayed in human islet cells. Diabetologia 2013;56:497-507. https://doi.org/10.1007/s00125-012-2779-7 |
||||
| 81 Petrenko V, Gandasi NR, Sage D, Tengholm A, Barg S, Dibner C: In pancreatic islets from type 2 diabetes patients, the dampened circadian oscillators lead to reduced insulin and glucagon exocytosis. Proc Natl Acad Sci U S A 2020;117:2484-2495. https://doi.org/10.1073/pnas.1916539117 |
||||
| 82 Petrenko V, Saini C, Giovannoni L, Gobet C, Sage D, Unser M, Heddad Masson M, Gu G, Bosco D, Gachon F, Philippe J, Dibner C: Pancreatic alpha- and beta-cellular clocks have distinct molecular properties and impact on islet hormone secretion and gene expression. Genes Dev 2017;31:383-398. https://doi.org/10.1101/gad.290379.116 |
||||
| 83 Schliess F, von Dahl S, Haussinger D: Insulin resistance induced by loop diuretics and hyperosmolarity in perfused rat liver. Biol Chem 2001;382:1063-1069. https://doi.org/10.1515/BC.2001.133 |
||||
| 84 Hallbrucker C, vom Dahl S, Lang F, Gerok W, Haussinger D: Modification of liver cell volume by insulin and glucagon. Pflugers Arch 1991;418:519-521. https://doi.org/10.1007/BF00497781 |
||||
| 85 Vom Dahl S, Hallbrucker C, Lang F, Gerok W, Haussinger D: Regulation of liver cell volume and proteolysis by glucagon and insulin. Biochem J 1991;278:771-777. https://doi.org/10.1042/bj2780771 |
||||
| 86 la Fleur SE, Kalsbeek A, Wortel J, Fekkes ML, Buijs RM: A daily rhythm in glucose tolerance: a role for the suprachiasmatic nucleus. Diabetes 2001;50:1237-1243. https://doi.org/10.2337/diabetes.50.6.1237 |
||||
| 87 Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA: BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol 2004;2:e377. https://doi.org/10.1371/journal.pbio.0020377 |
||||
| 88 Webster CR, Srinivasulu U, Ananthanarayanan M, Suchy FJ, Anwer MS: Protein kinase B/Akt mediates cAMP- and cell swelling-stimulated Na+/taurocholate cotransport and Ntcp translocation. J Biol Chem 2002;277:28578-28583. https://doi.org/10.1074/jbc.M201937200 |
||||
| 89 Lan WZ, Wang PY, Hill CE: Modulation of hepatocellular swelling-activated K+ currents by phosphoinositide pathway-dependent protein kinase C. Am J Physiol Cell Physiol 2006;291:C93-103. https://doi.org/10.1152/ajpcell.00602.2005 |
||||
| 90 Mora A, Lipina C, Tronche F, Sutherland C, Alessi DR: Deficiency of PDK1 in liver results in glucose intolerance, impairment of insulin-regulated gene expression and liver failure. Biochem J 2005;385:639-648. https://doi.org/10.1042/BJ20041782 |
||||
| 91 Liu H, Yu J, Xia T, Xiao Y, Zhang Q, Liu B, Guo Y, Deng J, Deng Y, Chen S, Naray-Fejes-Toth A, Fejes-Toth G, Guo F: Hepatic serum- and glucocorticoid-regulated protein kinase 1 (SGK1) regulates insulin sensitivity in mice via extracellular-signal-regulated kinase 1/2 (ERK1/2). Biochem J 2014;464:281-289. https://doi.org/10.1042/BJ20141005 |
||||
| 92 Schliess F, Haussinger D: Call volume and insulin signaling. Int Rev Cytol 2003;225:187-228. https://doi.org/10.1016/S0074-7696(05)25005-2 |
||||
| 93 Schliess F, Richter L, vom Dahl S, Haussinger D: Cell hydration and mTOR-dependent signalling. Acta Physiol (Oxf) 2006;187:223-229. https://doi.org/10.1111/j.1748-1716.2006.01547.x |
||||
| 94 Liu GY, Sabatini DM: mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 2020;21:183-203. https://doi.org/10.1038/s41580-019-0199-y |
||||
| 95 Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Ruegg MA, Hall MN: Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab 2012;15:725-738. https://doi.org/10.1016/j.cmet.2012.03.015 |
||||
| 96 Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM: mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 2010;468:1100-1104. https://doi.org/10.1038/nature09584 |
||||
| 97 Cornu M, Oppliger W, Albert V, Robitaille AM, Trapani F, Quagliata L, Fuhrer T, Sauer U, Terracciano L, Hall MN: Hepatic mTORC1 controls locomotor activity, body temperature, and lipid metabolism through FGF21. Proc Natl Acad Sci U S A 2014;111:11592-11599. https://doi.org/10.1073/pnas.1412047111 |
||||
| 98 Khapre RV, Kondratova AA, Patel S, Dubrovsky Y, Wrobel M, Antoch MP, Kondratov RV: BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging (Albany NY) 2014;6:48-57. https://doi.org/10.18632/aging.100633 |
||||
| 99 Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Guttler T, Davis F, Asara JM, Sahin M: The Circadian Protein BMAL1 Regulates Translation in Response to S6K1-Mediated Phosphorylation. Cell 2015;161:1138-1151. https://doi.org/10.1016/j.cell.2015.04.002 |
||||
| 100 Wu R, Dang F, Li P, Wang P, Xu Q, Liu Z, Li Y, Wu Y, Chen Y, Liu Y: The Circadian Protein Period2 Suppresses mTORC1 Activity via Recruiting Tsc1 to mTORC1 Complex. Cell Metab 2019;29:653-667.e6. https://doi.org/10.1016/j.cmet.2018.11.006 |
||||
| 101 Nakamura K, Brown RA, Narayanan S, Collins DL, Arnold DL, Alzheimer's Disease Neuroimaging I: Diurnal fluctuations in brain volume: Statistical analyses of MRI from large populations. Neuroimage 2015;118:126-132. https://doi.org/10.1016/j.neuroimage.2015.05.077 |
||||
| 102 Thomas C, Sadeghi N, Nayak A, Trefler A, Sarlls J, Baker CI, Pierpaoli C: Impact of time-of-day on diffusivity measures of brain tissue derived from diffusion tensor imaging. Neuroimage 2018;173:25-34. https://doi.org/10.1016/j.neuroimage.2018.02.026 |
||||
| 103 Hofman MA, Swaab DF: Diurnal and seasonal rhythms of neuronal activity in the suprachiasmatic nucleus of humans. J Biol Rhythms 1993;8:283-295. https://doi.org/10.1177/074873049300800402 |
||||
| 104 Usui S, Ikuno Y, Akiba M, Maruko I, Sekiryu T, Nishida K, Iida T: Circadian changes in subfoveal choroidal thickness and the relationship with circulatory factors in healthy subjects. Invest Ophthalmol Vis Sci 2012;53:2300-2307. https://doi.org/10.1167/iovs.11-8383 |
||||
| 105 Ashraf H, Nowroozzadeh MH: Diurnal variation of retinal thickness in healthy subjects. Optom Vis Sci 2014;91:615-623. https://doi.org/10.1097/OPX.0000000000000269 |
||||
| 106 Silagy C, Shelby-James T, Sage M, Wallage A: Patient-detected diurnal changes in spleen volume. Lancet 1998;352:710. https://doi.org/10.1016/S0140-6736(05)60828-8 |
||||
| 107 Watanabe M, Uchiyama Y: Twenty-four hour variations in subcellular structures of rat pancreatic islet B-, A- and D-cells, and of portal plasma glucose and insulin levels. Cell Tissue Res 1988;253:337-345. https://doi.org/10.1007/BF00222290 |
||||
| 108 Tomassen S, de Jonge HR, Tilly BC: Cell volume regulation in intestinal epithelial cells. Adv Exp Med Biol 2004;559:339-347. https://doi.org/10.1007/0-387-23752-6_31 |
||||