Corresponding Author: Amit Mishra
Cellular and Molecular Neurobiology Unit, Indian Institute of Technology Jodhpur, Jodhpur, Rajasthan 342037 (India)
Tel. +91-291-2801206 , E-Mail amit@iitj.ac.in
Resveratrol Promotes LRSAM1 E3 Ubiquitin Ligase-Dependent Degradation of Misfolded Proteins Linked with Neurodegeneration
Ankur Rakesh Dubeya Ribhav Mishraa Naveen Sundariaa Yuvraj Anandrao Jagtapa Prashant Kumara Sumit Kingera Akash Choudharya Hem Chandra Jhab Amit Prasadc Ravi Kumar Guttid Amit Mishraa
aCellular and Molecular Neurobiology Unit, Indian Institute of Technology Jodhpur, Jodhpur, Rajasthan, India, bInfection Bioengineering Group, Department of Biosciences and Biomedical Engineering, Indian Institute of Technology Indore, Indore, Madhya Pradesh, India, cSchool of Basic Sciences, Indian Institute of Technology Mandi, Mandi, Himachal Pradesh, India, dDepartment of Biochemistry, F49 School of Life Sciences University of Hyderabad, Hyderabad, Telangana, India
Introduction
Proteins are imperative for normal cellular functions and survival. The regular maintenance of protein turnover mechanisms fulfills the routine course of assigned cellular functions to achieve proteostasis [1, 2]. Mutant or abnormal protein accumulation can trigger an alarm to quickly eliminate their aggregation from different cellular compartments. Failure in the degradation mechanisms of aggregation-prone proteins can cause critical neurodegenerative diseases and develop clinical symptoms of imperfect ageing [3, 4]. The removal of misfolded proteins from the different cellular compartments adds an extra metabolic burden on the cell. Under such circumstances, cells initially prefer their folding, utilizing molecular chaperones, prior to designating them for degradation [5]. Depletion in the chaperone capacity promotes the occurrence of neurodegeneration and ageing [6]. However, a complete understanding of protein folding mechanisms is still truant, and therefore it is challenging to come up with molecular strategies that can assist in improving chaperone functioning for better folding of aberrant proteins. Furthermore, the molecular mechanisms by which cells can specifically target the removal of abnormal or poorly folded proteins from the intracellular pool are unknown. It is also important to find out strategies based on small molecules that can improve the cellular PQC mechanisms for the effective clearance of damaged proteins to maintain overall proteostasis.
Cells continuously monitor the folding mechanism of damaged proteins; dysfunction in protein folding activates cellular proteolytic mechanisms [6-8]. Ubiquitin Proteasome System (UPS) is the primary selective proteolytic process in cells that removes aberrant client proteins [9]. Cells can recognize and separate the aberrant proteins for their removal with the help of E3 ubiquitin ligase enzymes of the UPS [10]. In cells, utilization of a few E3 ubiquitin ligases that specifically participate in cellular PQC mechanisms generates the first line of cellular defense against proteotoxic insults [11, 12]. Previous findings suggest that mutations or dysfunctions of E3 ubiquitin ligases contribute to neurodegenerative and neurodevelopmental disorders [13-15]. In cells under stress conditions, altered functions of quality control E3 ubiquitin ligases contribute to the proteostasis imbalance leading to proteotoxic insults [10].
Depleted molecular functions of PQC-E3 ubiquitin ligases consequently result into the build-up of aggregation-prone proteins affecting cellular health [16]. Over accumulation of misfolded protein inclusions can inhibit the proteasome system and may activate the autophagic pathway for the removal of unwanted aggresomes [17, 18]. Such findings suggest that upregulation of PQC-E3 ubiquitin ligases may ameliorate proteotoxicity and support the re-establishment of healthy proteostasis. However, the major challenge is to find out the molecular strategy by which we can activate or upregulate the functions of perturbed PQC-E3 ubiquitin ligases against aggregation-prone proteins associated with cellular damage.
Earlier studies have suggested that few natural compounds demonstrate strong anti-aggregation capabilities, and the use of such compounds may suppress the accumulation of proteins, which is likely an approach to minimize cellular damages [19-22]. Resveratrol is found in raspberries and grapes, belonging to stilbene compounds. Interestingly, dietary supplement consumption of resveratrol decreases plaques synthesis in an animal model of Alzheimer’s disease [23, 24]. Moreover, resveratrol treatment in early Parkinson’s disease patients demonstrates improved chaperone networks along with enhanced protein degradation and redox balance restoring cytoprotection in their skin fibroblasts [25]. Such findings gave us a clue that resveratrol likely exhibits an inductive potential to improve the functions of PQC mechanisms against the abnormal proteinaceous inclusions. But, still, the mechanism by which resveratrol reduces the overburden of misfolded protein accumulation and destabilizes amorphous protein aggregates is unknown.
In our present investigation, we found that exposure of resveratrol elevates the endogenous levels of LRSAM1 E3 ubiquitin ligase as well as slows down its intracellular turnover; earlier, it has been reported that mutations in LRSAM1 cause spongiform neurodegeneration [26] and it also exhibits PQC competence, which suppresses misfolded proteins aggregation [10]. We observed that resveratrol treatment efficiently inhibits aggregation of bona fide mutant proteins and also encourages the clearance of thermally misfolded luciferase inclusions. Our current findings imply that the use of resveratrol may offer promising cytoprotective molecular strategies, which can alter the functions of PQC-components that, consequently induces the degradation of aberrant proteins. Altogether our results suggest that most likely, in the future we can develop a molecular strategy to prevent the loss of proteostasis via the removal of over accumulated toxic aggregates to overcome the impairment of PQC mechanisms.
Materials and Methods
Materials
Resveratrol, chloroquine, cycloheximide, 2-mercaptoethanol (BME), lactacystin, MG132, TRIzol, and all reagents for cell culture were procured from Sigma. OptiMEM and Lipofectamine® 2000 were acquired from Life Technologies. Luciferase antibody, anti-ubiquitin, anti-LRSAM1, anti-β actin, anti-GAPDH, anti-c-myc, and anti-actin antibodies were purchased from Santa Cruz Biotechnology. Anti-Lamp2 antibody was obtained from Thermo Fisher Scientific, anti-LC3 antibody was procured from Pierce antibodies, anti-p62/SQSTM1 antibody was procured from Sigma, and LRSAM1 antibody was obtained from Cell Signaling Technology. iScriptTM One-Step RT-PCR Kit along with SYBR® was procured from Bio-Rad Laboratories. Proteasome-GloTM assay reagents and Dual-luciferase reporter gene assay kits were obtained from Promega. Fluorescein isothiocyanate-conjugated (FITC) IgG anti-mouse and anti-rabbit, Rhodamine-conjugated IgG anti-mouse and anti-rabbit and horseradish peroxidase (HRP) conjugated IgG anti-mouse and anti-rabbit antibodies were acquired from Vector Laboratories. Luciferase-pcDNA3 (Addgene 18964) and pcDNA3-cMyc (Addgene 16011) were procured from Addgene, pcDNA™ 3.1 was obtained from Life Technologies, LRSAM1ΔRING-Myc was synthesized by digesting LRSAM1-Myc plasmid with BglII for 3 hours at 37°C and further T4 DNA ligase mediated self-ligation was performed overnight at 37°C; LRSAM1-specific siRNA oligonucleotides were obtained from Life Technologies.
Cell Culture Experiments
A549 and COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) comprising 10% fetal bovine serum (heat-inactivated) along with streptomycin (100 µg/ml)/penicillin (100 U/ml) antibiotic. Just before transfection, different culture plates were seeded with a sub-confluent density of cells. Transient transfection with different plasmid constructs was achieved using Lipofectamine® 2000 reagent following the instructions provided by the manufacturer. Post 24 h, cells were exposed to different chemical molecules and were prepared for immunoblotting or immunofluorescence staining. To assess the influence of resveratrol on cell morphology, COS-7 cells were subjected to different concentrations of resveratrol for diverse time intervals and visualized under a bright-field microscope.
Immunofluorescence Technique
COS-7 cells were plated into the 2-well chamber slides and then briefly transfected with different constructs and treated with control or resveratrol or MG132 or lactacystin. Post this, the slides were treated with phosphate-buffered saline (PBS) thrice, and fixation with 4% paraformaldehyde solution (PBS) for 15-20 minutes was done. They were then treated with 0.5% Triton X-100 for cell membrane permeabilization (5 mins), thoroughly washed with PBS for 3-4 times, and incubated in 2% rabbit serum solution (PBS) for a minimum of 30 mins. Post this, the cells were incubated overnight with different primary antibody (1:500 dilution) at 4°C, and afterward thoroughly washed with PBS for three times (5 mins each) and treated with secondary antibody conjugated either with rhodamine or fluorescein isothiocyanate for 1-4 hours. Afterward, the cells were mounted with an antifade solution and 4’,6-diamidino-2-phenylindole (DAPI) to stain nuclei as described earlier [27]. Then the prepared slides were observed under a fluorescence microscope.
Immunoblotting Technique
Transient transfection was performed with different plasmids on the cells maintained in 6-well plates and treated with control or resveratrol or MG132. Post exposure, whole cell lysates were made and used for performing SDS-PAGE (Bio-Rad Laboratories). Electrophoresed samples were transported onto the nitrocellulose membrane. Blocking was done using 5% non-fat milk in Tris-buffered saline Tween 20 (TBST) [50 mM Tris; 0.15 M NaCl, 0.1% Tween 20, pH 7.4] for not less than an hour, then were treated overnight at 4°C with suitable primary antibody solution in TBST (1:1000 dilution). After thoroughly washing with TBST for 3-4 times (5 mins each), the nitrocellulose membrane was treated with a secondary antibody conjugated with HRP enzyme (1:10000) for not less than 30 minutes. Development of blots was then carried out with Luminata Crescendo Western HRP substrate (EMD Millipore).
Cycloheximide Chase Experiment and Reporter Gene Assay
Resveratrol or control-treated COS-7 cells into 6-well plates were further treated with cycloheximide (15 µg/ml) for multiple time points. Total protein isolation was done, and the immunoblotting experiment was performed with anti-LRSAM1 and anti-GAPDH primary antibodies. 6-well plates having COS-7 were used for transient transfection with LRSAM1-Myc and luciferase plasmids; some wells were subjected to either MG132 or chloroquine or resveratrol. Post this, the cells were kept for 30 minutes at 43°C and re-incubated under general incubation environment for 2 hours and then advanced to immunoblotting experiment with anti-luciferase and anti-β-Actin antibodies. Similar experimental setup was utilized for analyzing luciferase activity via using Dual-luciferase reporter gene assay kit (Promega) following the maker’s protocol.
Reverse Transcriptase-Polymerase Chain Reaction Analysis (RT-PCR)
A549 cells (primer compatible) were maintained onto 6-well culture plates and subjected to either control or resveratrol with different concentrations and different periods. Post-treatment, cells were harvested to isolate whole-cell RNA with the help of TRIzol. Further RT-PCR was performed for analyzing LRSAM1 and β-actin using the primer sequence; LRSAM1F, 5’-AACGCCTGGAGTACCAGATG-3’, LRSAM1R, 5’-AACCTTGATGGTTGCCAGAC-3’, β-actinF, 5’-ATCGTCCACCGCAAATGCTTCTA-3’, β-actinR, 5’-AGCCATGCCAATCTCATCTTGTT-3’. The complementary DNA (cDNA) was first synthesized utilizing the total cell RNA with the help of iScriptTM cDNA synthesis kit (Bio-Rad Laboratories) and amplified using SsoFastTM EvaGreen® Supermix and MyiQTM Two-Color Real-Time PCR Detection System (Bio-Rad Laboratories). The parameters for carrying out 37 cycles of PCR with LRSAM1 primers were 94°C 30s, 55°C 30s, 72°C 30s and 72°C 2 min (final extension). The reaction conditions for 25 cycles with β-actin primers were 94°C 30s, 55°C 45s, 72°C 30s, and 72°C 2 min (final extension).
Proteasome Activity Assay and Aggregate Counting
COS-7 cells were transiently transfected by LRSAM1-Myc or LRSAM1ΔRING-Myc or LRSAM1-siRNA and utilized to carry out proteasome activity assay with Proteasome-GloTM systems (Promega) based on the protocol of the manufacturer. All the experiments were done in triplicates to analyze changes in proteasome activity. Brief transfection of COS-7 cells was performed for 48 h with pd1EGFP, followed by control or MG132 or resveratrol treatment to quantify misfolded d1EGFP aggregates. Cells with greater than one aggresome were marked as one prominent inclusion, and fluorescence microscopy (~450 transfected cells in both cases) was utilized to visualize and quantify the misfolded d1EGFP aggregates.
Results
Effects of Resveratrol on the Endogenous Level of E3 Ubiquitin Ligase LRSAM1 Linked With Cellular Protein Quality Control Mechanism
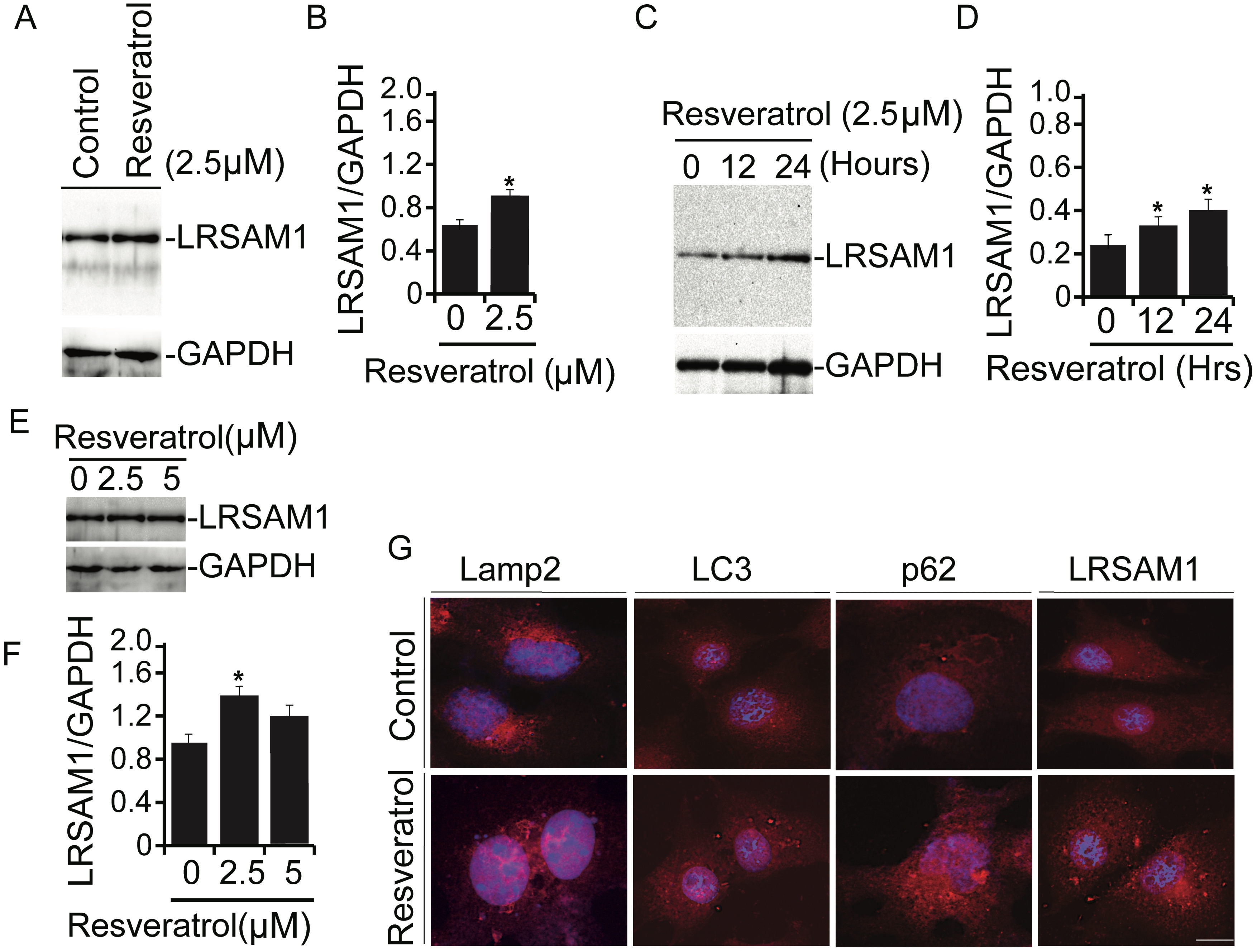
Polyphenol resveratrol, a natural compound, retains antioxidant properties. To elucidate the molecular characteristics of resveratrol against abnormal proteins, we conducted the analysis of cellular PQC components following resveratrol treatment. As shown in Fig. 1A-1B, we surprisingly observed that LRSAM1 endogenous level was increased after exposure of cells to resveratrol. Preliminary results suggested that treatment of resveratrol induces LRSAM1 levels. To further confirm these findings, similar experiments were performed at different time periods (Fig. 1C-1D) and in a concentration-dependent (Fig. 1E-1F) manner [28] and noticed similar effects of resveratrol as observed in the earlier analysis.
The elevated levels of LRSAM1 in response to resveratrol treatment suggest the relevant anti-aggregation molecular cope-up strategy of cells against intractable misfolded protein aggregates. To investigate the cytoprotective prospect of resveratrol over abnormal proteins via PQC components modulation, we performed an immunofluorescence analysis with or without resveratrol. As shown in Fig. 1G micrographs, exposure of resveratrol marginally upregulated various PQC components such as Lamp2, LC3, and p62 proteins. But in a similar experiment, treatment of resveratrol significantly induced the endogenous levels of LRSAM1 E3 ubiquitin ligase. These results indicate that resveratrol treatment can affect the levels of PQC components up to a considerable extent, which may generate cytoprotection against proteotoxic insults.
Expression of LRSAM1 Can Elevate Intracellular Proteasome Activities
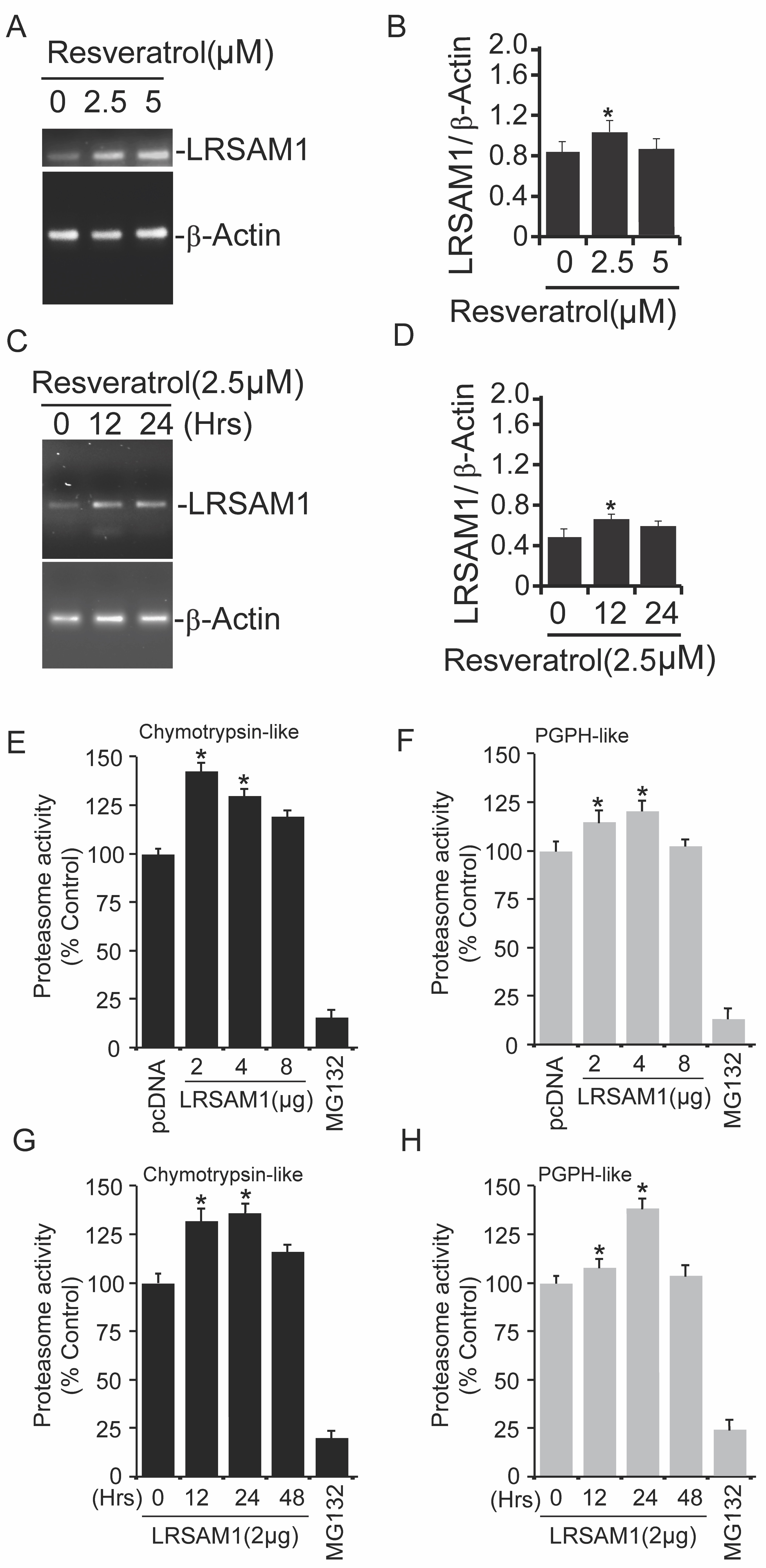
Previous findings suggest that resveratrol induces proteasomal degradation of Nanog via p53 activation [29]. In our current findings, we have noticed the elevation of LRSAM1 mRNA levels after resveratrol treatment in both concentration (Fig. 2A-2B) as well as time-dependent (Fig. 2C-2D) manner. Our earlier study demonstrates that LRSAM1 can target misfolded proteins for their intracellular degradation [10]. These findings prompt us to further characterize the detailed modulatory effects of LRSAM1 E3 ubiquitin ligase on intracellular protein destruction mechanism. Therefore, we have briefly transfected COS-7 cells with wild-type LRSAM1 plasmids in different concentration profiles. Post-transfected cell extracts were used for proteasome assay as described in the methods section.
We have observed elevation in both chymotrypsin-like (Fig. 2E) as well as post-glutamyl peptide hydrolase (PGPH)-like (Fig. 2F) protease activities of the proteasome after transient transfection of LRSAM1 construct at various concentrations. In a similar way, to further confirm the effect of LRSAM1 at different time intervals, we next performed other sets of proteasome assays with both chymotrypsin-like (Fig. 2G) PGPH-like protease activities (Fig. 2H), exogenous expression of LRSAM1 also induced the protease activities of proteasome at different time points. These results suggest that the upregulation/overexpression of LRSAM1 can modulate proteasome activities at varying concentrations and times.
Inactive Form of LRSAM1 Did Not Induce Proteasome Activities
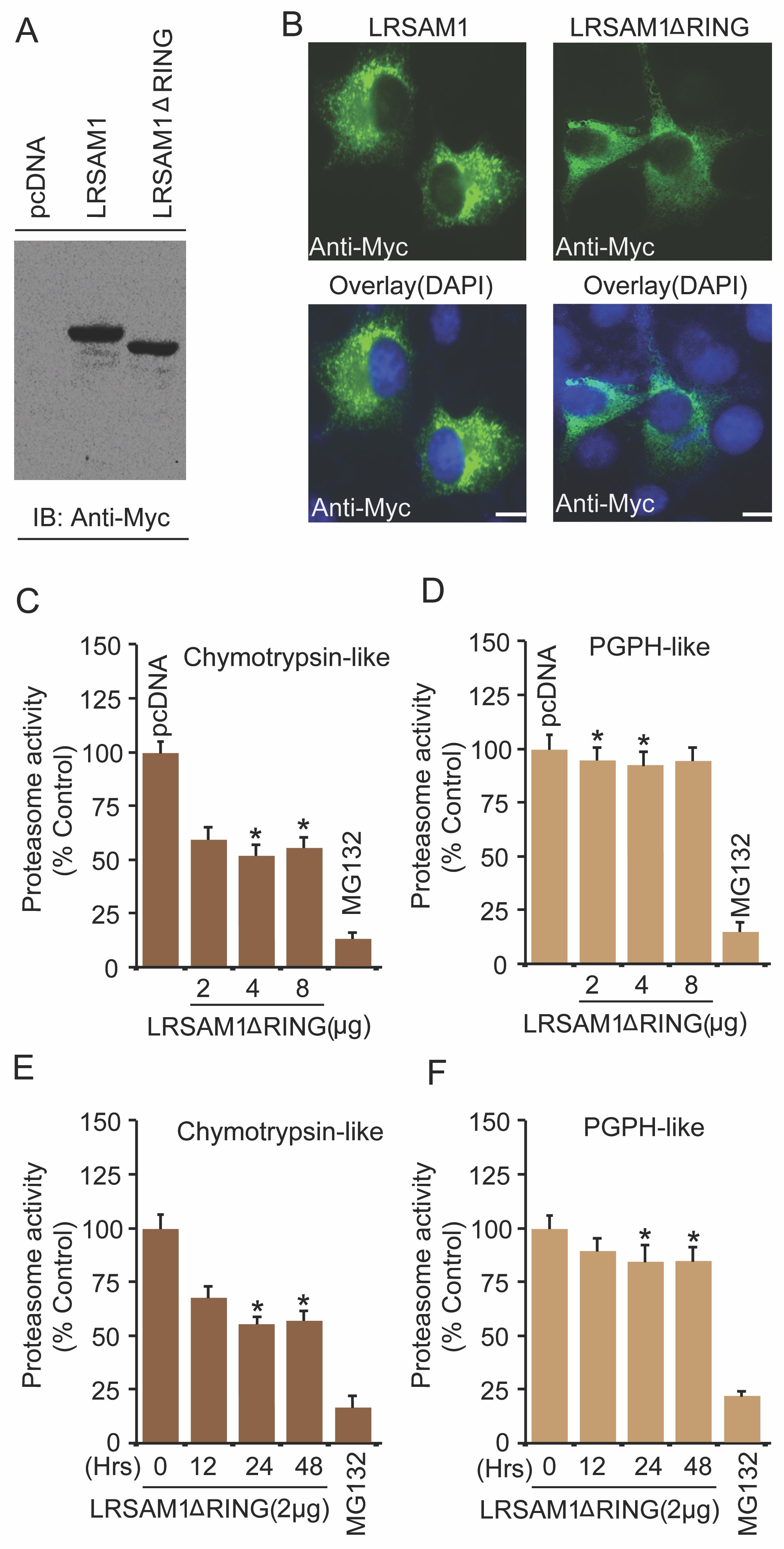
Several intracellular regulatory systems respond to abnormal protein aggregation problems. Certain E3 ubiquitin ligases function specifically for PQC and finally target aberrant proteins for clearance. Previously, our study suggests that LRSAM1 also recognizes misfolded proteins for their degradation [10]. To further validate the function of LRSAM1 E3 ubiquitin ligase in moderating the proteasome function, cells were briefly transfected with LRSAM1ΔRING mutant, the catalytically inactive form of the full-length LRSAM1 construct. The expression of catalytically inactive LRSAM1ΔRING mutant was confirmed by both immunoblotting and immunofluorescence analysis, as shown in Fig. 3A and 3B, respectively. We then examined the effects of LRSAM1ΔRING on both chymotrypsin-like and PGPH-like protease activities. Next, we detected that exogenous expression of LRSAM1ΔRING mutant suppressed both proteasome chymotrypsin-like (Fig. 3C) and PGPH-like protease (Fig. 3D) activities in a concentration-dependent fashion. A similar effect of LRSAM1ΔRING mutant was found on the above-described proteasome chymotrypsin-like (Fig. 3E) and PGPH-like protease (Fig. 3F) activities when cells were transiently transfected at various time intervals.
Depletion of LRSAM1 Cannot Induce Proteasome Activities
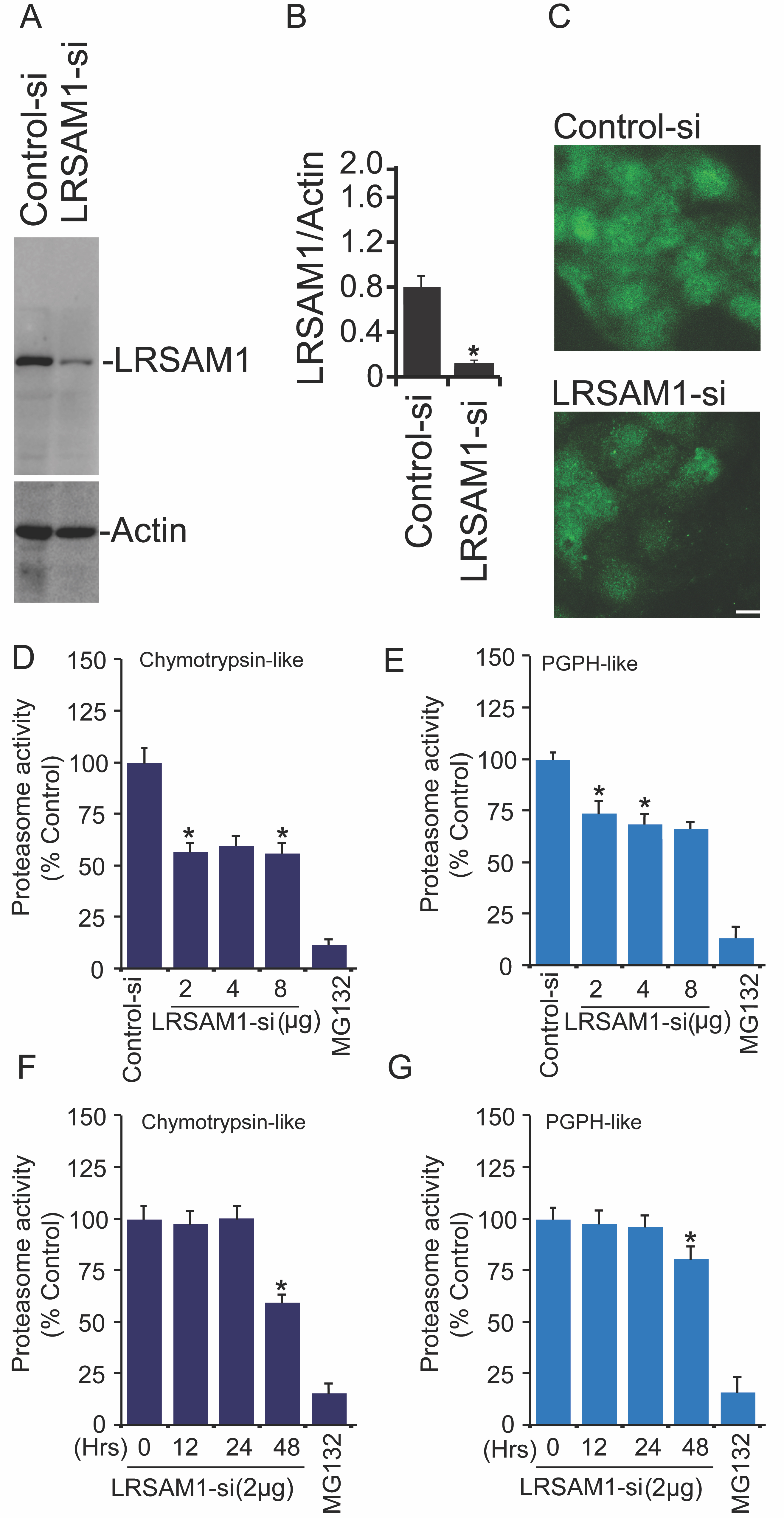
Because we have observed the induction of proteasome activities after exogenous expression of normal LRSAM1 in comparison to inactive LRSAM1ΔRING form, it encouraged us to examine the consequence of LRSAM1 depletion on various proteasome activities. To test this question, we knock down the endogenous levels of LRSAM1 by using the siRNA transfection approach. Transient transfection of A549 cells with control and LRSAM1-siRNA was performed and post-transfected cell extracts were used for the verification of siRNA-mediated LRSAM1 depletion via immunoblotting (Fig. 4A-4B) and immunofluorescence analysis as depicted in Fig. 4C micrographs. Next, to address the above question, we performed detailed proteasome assays in the presence of different concentrations of LRSAM1-siRNA. Proteasome assay analysis revealed that both chymotrypsin-like (Fig. 4D) and PGPH-like (Fig. 4E) protease activities of proteasome were suppressed when LRSAM1 was partially depleted from the cells. Similar results were obtained when LRSAM1-siRNA were transfected at different time intervals, and post-transfected cells were utilized for measuring chymotrypsin-like (Fig. 4F) as well as PGPH-like (Fig. 4G) protease activities. Altogether, this set of results shows that depletion of LRSAM1 can marginally suppress proteasome activities.
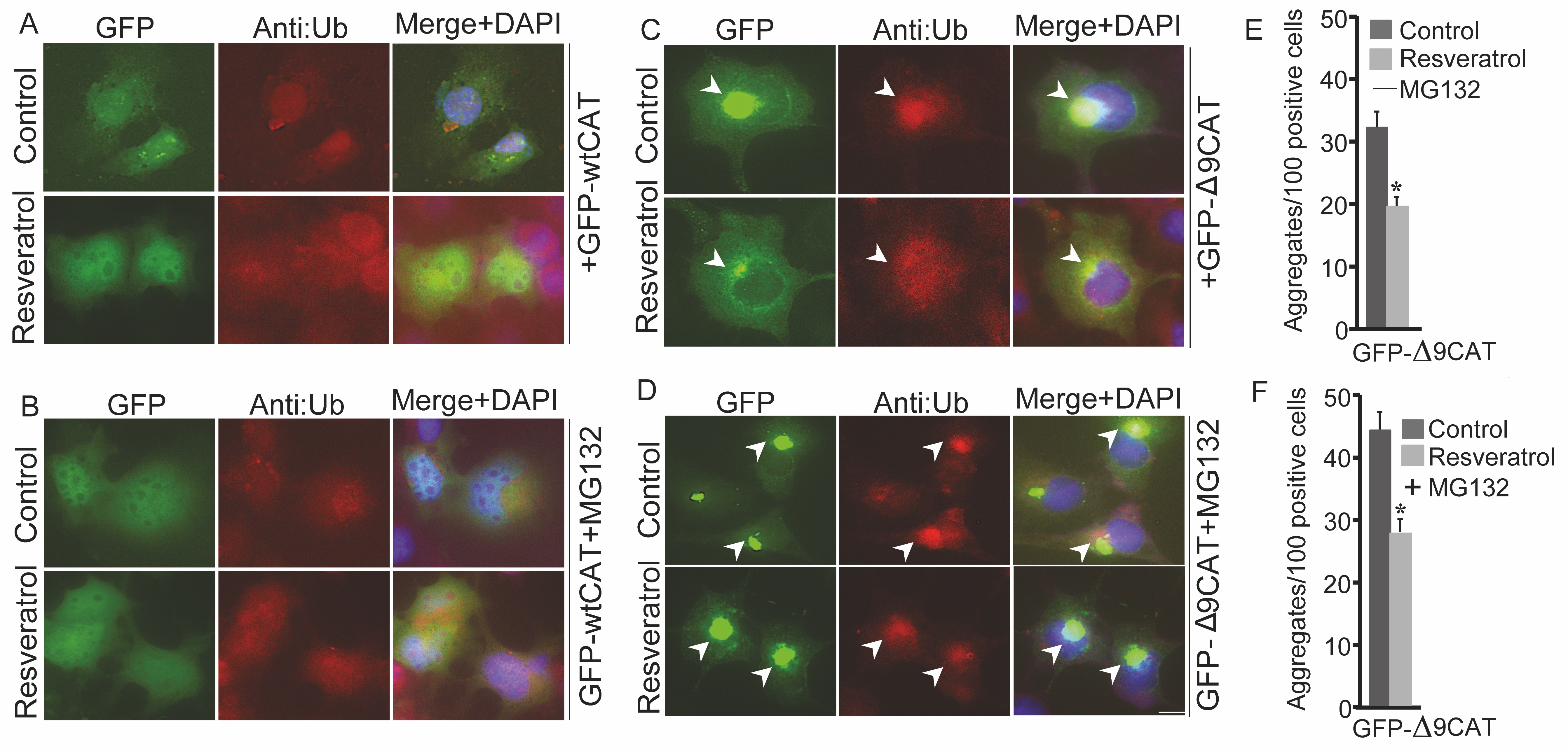
Resveratrol Treatment Reduces the Aggregation of Aberrant Proteins
Several neurodegenerative diseases are a result of the over-build-up of aberrant proteins. Thus, it is crucial to check the outcome of resveratrol exposure on aggregation propensity of mutant bona fide misfolded proteins. COS-7 cells were transitorily transfected with GFP-wtCAT (Fig. 5A and 5B) and GFP-Δ9CAT (Fig. 5C and 5D) constructs. Certain sets of post-transfected cells were exposed to resveratrol and putative proteasome inhibitor MG132 (Fig. 5A-5D). Exogenous GFP-Δ9CAT expression produced intracellular aggregates in comparison to GFP-wtCAT protein. Exposure of resveratrol dramatically reduced the number as well as the size of misfolded GFP-Δ9CAT protein aggregates (Fig. 5C and 5E); the addition of MG132 resulted in further accumulation of distinct perinuclear GFP-Δ9CAT aggresomes (Fig. 5D and 5F). Consistent with our preliminary findings, here we observed that resveratrol treatment induces the clearance of aberrant protein accumulation, probably via influencing the function of cellular PQC machinery and its molecular components.
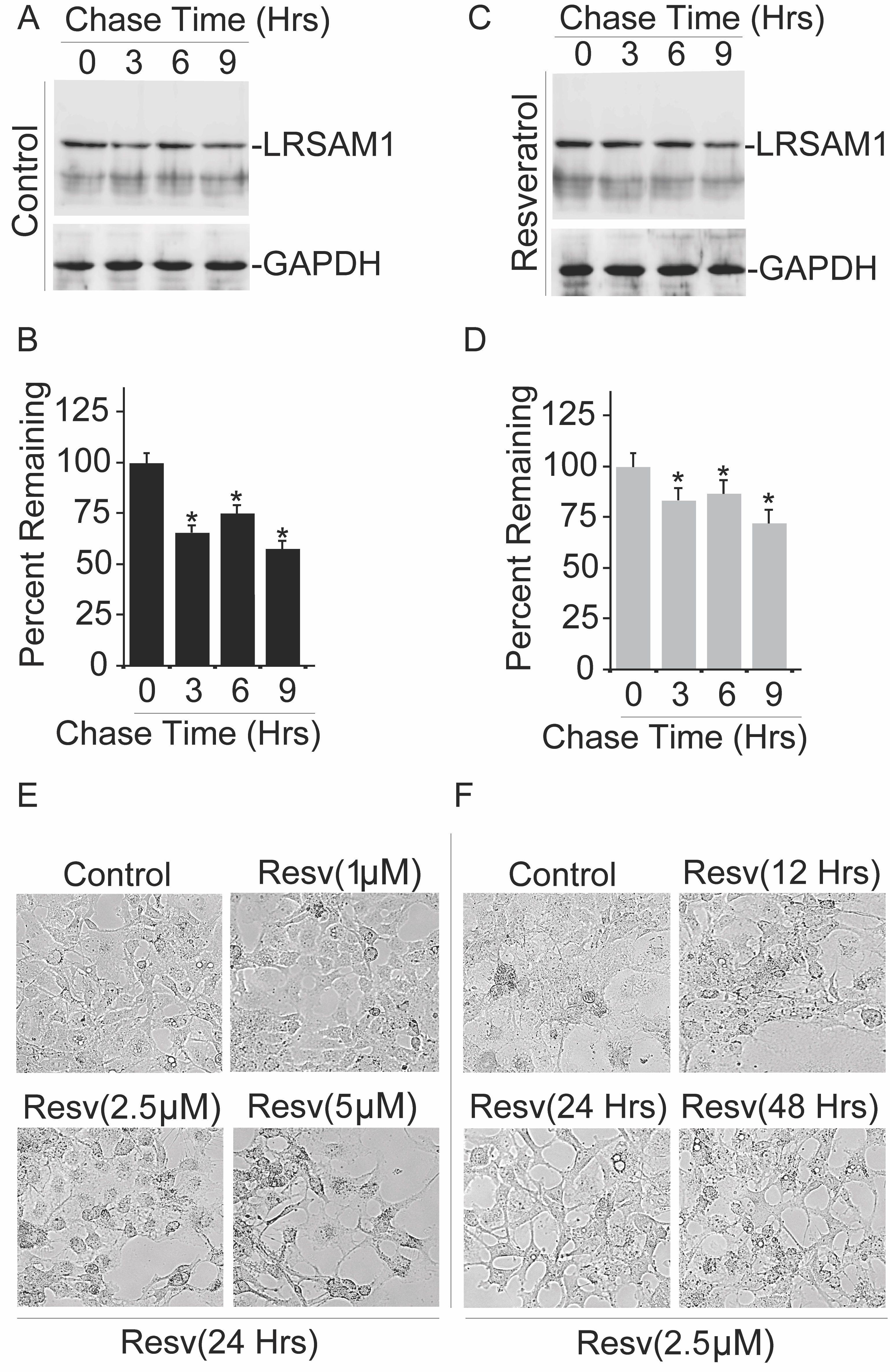
Resveratrol Stabilizes Endogenous Level of LRSAM1 Protein
To determine how the treatment of resveratrol influences the intracellular amounts of LRSAM1 E3 ubiquitin ligase, we determined whether the half-life of LRSAM1 is affected in the presence of resveratrol. We carried out a cycloheximide chase analysis to quantify the half-life of LRSAM1, with or without resveratrol (Fig. 6A-6D). In the chase experiment, synthesis of new proteins had been blocked by cycloheximide; collected cell lysates of chase-experiment at different time points were used for immunoblotting analysis via using LRSAM1 antibody. As shown in Fig. 6C and 6D, protein amounts of LRSAM1 were significantly stabilized after the treatment of resveratrol as compared to the control experiment (Fig. 6A and 6B). Simultaneously, it is important for us to determine whether resveratrol addition affects overall cellular viability or not; therefore, cells were subjected to resveratrol treatment at different concentrations and time intervals, and bright-field images were acquired as shown in Fig. 6E and 6F, respectively. In our current observation, no significant adverse effects of resveratrol were noticed on cell viability. Notably, the turnover rate of LRASM1 protein was more stabilized when exposed to resveratrol, suggesting that the exposure of resveratrol to the LRSAM1 protein turnover is specific and does not affect cell viability.
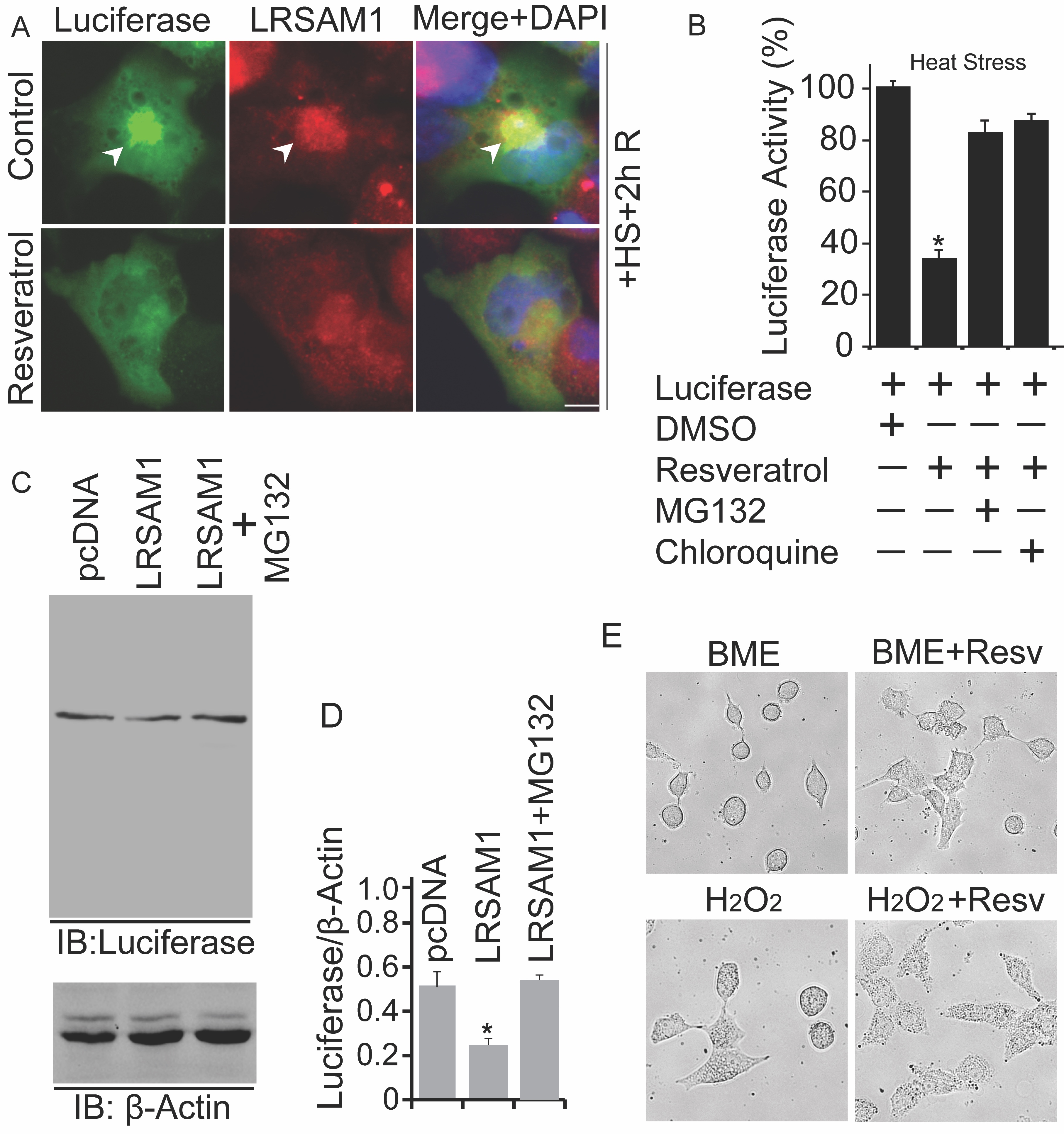
Resveratrol Promotes Heat-Denatured Luciferase Protein Degradation and Protects Against Cellular Stresses
Proteotoxic insults associated with cell death are majorly caused by the build-up of misfolded proteins. Despite this, we know that resveratrol may retain the molecular capabilities to improve the functions of PQC mechanism, but it is critical for us to detect the outcome of resveratrol exposure on the levels of various model-misfolded proteins. In order to examine this, cells were briefly transfected with a luciferase-containing plasmid construct. Post-transfected cells were subjected to 43°C for 30 minutes, and recovery was attained at 37°C for 02 hours. Similarly, some cells were treated with resveratrol and used for immunofluorescence analysis using anti-LRSAM1 antibody (Fig. 7A). Treatment of resveratrol elevated the cellular levels of LRSAM1 and promotes the degradation of aberrant heat-distorted luciferase inclusions. As described above, for luciferase assay, identical sets of cells were used (Fig. 7B). Furthermore, compared to the control (DMSO), the addition of resveratrol induces the clearance of thermally-misfolded luciferase, and the effect of resveratrol was suppressed by the treatment of MG132 and chloroquine. Immunoblot analysis (Fig. 7C and 7D) also confirmed that exogenous expression of LRSAM1 promotes the clearance of heat-denatured proteins and was stopped by MG132 addition. Accumulation of misfolded proteins can cause various types of stress, such as oxidative and ER stress conditions [26]. As demonstrated in Fig. 7E, treatment of resveratrol possesses a cytoprotective response against ER (2-Mercaptoethanol; BME) and oxidative stress (H2O2) inducing agents.
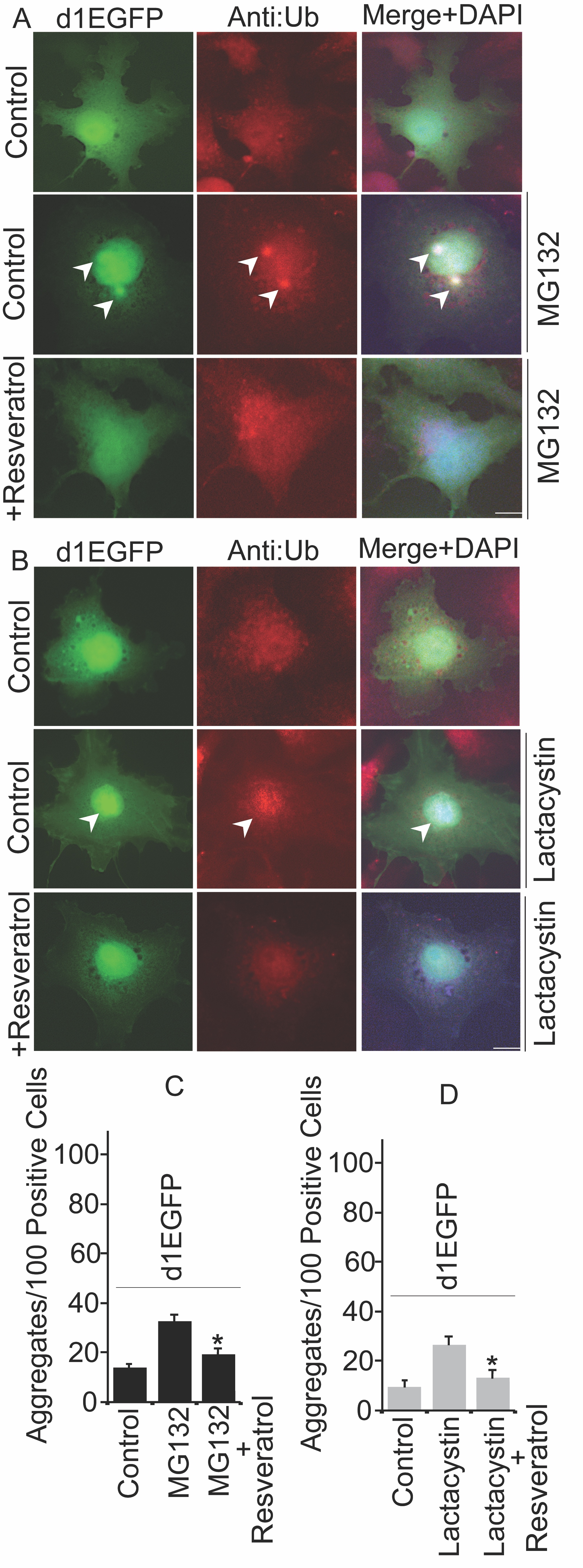
Resveratrol-Based PQC Moderation Capabilities Alleviates Cytotoxicity
Polyubiquitinated proteins are generally degraded by proteasome machinery. Inhibition of proteasome may lead to the build-up of aberrant proteins, causing proteotoxic insults in cells [30]. To test the notion of how resveratrol can affect overall aggregation propensity, we transiently transfected a model proteasome substrate pd1EGFP plasmid that retains proline, glutamic acid, serine, and threonine signal sequence that can be used for specifically targeting for proteasome degradation. As shown in Fig. 8A and 8C, inhibition of proteasome by MG132 forms ubiquitinated d1EGFP inclusions, and the addition of resveratrol reduces the formation of those abnormal inclusions. To further confirm the above results, next, we used lactacystin, another putative proteasome inhibitor; as demonstrated in Fig. 8B and 8D, treatment of lactacystin induces the abundance of d1EGFP aggresomes, which can be reduced by the exposure of resveratrol. Because treatment of resveratrol decreases the number of aggregates, we thought to use this as a reliable strategy to further validate the cytoprotective nature of resveratrol based on the elevation of LRSAM1 levels. Collectively, our results suggest that resveratrol exhibits cytoprotective PQC moderation capabilities, and most likely; it targets LRSAM1 E3 ubiquitin ligase for clearing over accumulated or misfolded proteins.
We are also greatly thankful for the gifted plasmids as described here: GFP-wtCAT and GFP-Δ9CAT: Dr. Csaba Soti (Department of Medical Chemistry, Semmelweis University, Budapest, Hungary), Luciferase-pcDNA3: Dr. William Kaelin from Dana Farber Cancer Institute and the Howard Hughes Medical Institute, LRSAM1-Myc: Dr. R. Burgess (The Jackson Laboratory, USA) and pd1EGFP: Dr. Nihar Ranjan Jana (National Brain Research Centre, Manesar, Gurgaon, India).
Author Contributions
A.R.D, R.M, N.S, Y.A.J, P.K, S.K and A.C performed the designed experiments and implemented the research. H.C.J, A.P and R.K.G verified the analytical methods and helped in generation of framework of experiments and perform critical analysis of manuscript. A.M overall design various experiments. Also contributed to the analysis of the results and perform the writing of the manuscript. All authors discussed the results and contributed to the final manuscript.
Funding
AM received BRNS grant 54/14/16/2020-BRNS from Government of India and also AM research grant EMR/2016/000716 from Science and Engineering Research Board (SERB), Department of Science & Technology, Government of India supported the research work.
The authors declare that no conflicts of interest exist.
| 1 Dörrbaum AR, Kochen L, Langer JD, Schuman EM: Local and global influences on protein turnover in neurons and glia. Elife 2018;7:e34202. https://doi.org/10.7554/eLife.34202 |
||||
| 2 Hipp MS, Kasturi P, Hartl FU: The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol 2019;20:421-435. https://doi.org/10.1038/s41580-019-0101-y |
||||
| 3 Romine IC, Wiseman RL: Starting at the beginning: endoplasmic reticulum proteostasis and systemic amyloid disease. Biochem J 2020;477:1721-1732. https://doi.org/10.1042/BCJ20190312 |
||||
| 4 Ruan L, Zhou C, Jin E, Kucharavy A, Zhang Y, Wen Z, Florens L, Li R: Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 2017;543:443-446. https://doi.org/10.1038/nature21695 |
||||
| 5 Mogk A, Bukau B, Kampinga HH: Cellular handling of protein aggregates by disaggregation machines. Mol Cell 2018;69:214-226. https://doi.org/10.1016/j.molcel.2018.01.004 |
||||
| 6 Feleciano DR, Juenemann K, Iburg M, Brás IC, Holmberg CI, Kirstein J: Crosstalk between chaperone-mediated protein disaggregation and proteolytic pathways in aging and disease. Front Aging Neurosci 2019;11:9. https://doi.org/10.3389/fnagi.2019.00009 |
||||
| 7 Díaz-Villanueva JF, Díaz-Molina R, García-González V: Protein folding and mechanisms of proteostasis. Int J Mol Sci 2015;16:17193-17230. https://doi.org/10.3390/ijms160817193 |
||||
| 8 Kevei É, Pokrzywa W, Hoppe T: Repair or destruction-an intimate liaison between ubiquitin ligases and molecular chaperones in proteostasis. FEBS Lett 2017;591:2616-2635. https://doi.org/10.1002/1873-3468.12750 |
||||
| 9 Pohl C, Dikic I: Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 2019;366:818-822. https://doi.org/10.1126/science.aax3769 |
||||
| 10 Mishra R, Amanullah A, Upadhyay A, Dhiman R, Dubey AR, Singh S, Prasad A, Mishra A: Ubiquitin ligase LRSAM1 suppresses neurodegenerative diseases linked aberrant proteins induced cell death. Int J Biochem Cell Biol 2020;120:105697. https://doi.org/10.1016/j.biocel.2020.105697 |
||||
| 11 Jayaraj GG, Hipp MS, Hartl FU: Functional modules of the proteostasis network. Cold Spring Harb Perspect Biol 2020;12:a033951. https://doi.org/10.1101/cshperspect.a033951 |
||||
| 12 Chhangani D, Joshi AP, Mishra A: E3 ubiquitin ligases in protein quality control mechanism. Mol Neurobiol 2012;45:571-585. https://doi.org/10.1007/s12035-012-8273-x |
||||
| 13 George AJ, Hoffiz YC, Charles AJ, Zhu Y, Mabb AM: A comprehensive atlas of E3 ubiquitin ligase mutations in neurological disorders. Front Genet 2018;9:29. https://doi.org/10.3389/fgene.2018.00029 |
||||
| 14 Upadhyay A, Joshi V, Amanullah A, Mishra R, Arora N, Prasad A, Mishra A: E3 ubiquitin ligases neurobiological mechanisms: development to degeneration. Front Mol Neurosci 2017;10:151. https://doi.org/10.3389/fnmol.2017.00151 |
||||
| 15 Upadhyay A, Amanullah A, Chhangani D, Mishra R, Prasad A, Mishra A: Mahogunin Ring Finger-1 (MGRN1), a Multifaceted Ubiquitin Ligase: Recent Unraveling of Neurobiological Mechanisms. Mol Neurobiol 2016;53:4484-4496. https://doi.org/10.1007/s12035-015-9379-8 |
||||
| 16 Kanack AJ, Newsom OJ, Scaglione KM: Most mutations that cause spinocerebellar ataxia autosomal recessive type 16 (SCAR16) destabilize the protein quality-control E3 ligase CHIP. J Biol Chem 2018;293:2735-2743. https://doi.org/10.1074/jbc.RA117.000477 |
||||
| 17 Johnston HE, Samant RS: Alternative systems for misfolded protein clearance: life beyond the proteasome. FEBS J 2020;288:4464-4487. https://doi.org/10.1111/febs.15617 |
||||
| 18 Thibaudeau TA, Anderson RT, Smith DM: A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat Commun 2018;9:1-14. https://doi.org/10.1038/s41467-018-03509-0 |
||||
| 19 Kumar S, Krishnakumar VG, Morya V, Gupta S, Datta B: Nanobiocatalyst facilitated aglycosidic quercetin as a potent inhibitor of tau protein aggregation. Int J Biol Macromol 2019;138:168-180. https://doi.org/10.1016/j.ijbiomac.2019.07.081 |
||||
| 20 Prajapati KP, Singh AP, Dubey K, Ansari M, Temgire M, Anand BG, Kar K: Myricetin inhibits amyloid fibril formation of globular proteins by stabilizing the native structures. Colloids Surf B Biointerfaces 2020;186:110640. https://doi.org/10.1016/j.colsurfb.2019.110640 |
||||
| 21 Ren B, Liu Y, Zhang Y, Cai Y, Gong X, Chang Y, Xu L, Zheng J: Genistein: a dual inhibitor of both amyloid β and human islet amylin peptides. ACS Chem Neurosci 2018;9:1215-1224. https://doi.org/10.1021/acschemneuro.8b00039 |
||||
| 22 Sonawane SK, Chidambaram H, Boral D, Gorantla NV, Balmik AA, Dangi A, Ramasamy S, Marelli UK, Chinnathambi S: EGCG impedes human Tau aggregation and interacts with Tau. Sci Rep 2020;10:1-17. https://doi.org/10.1038/s41598-020-69429-6 |
||||
| 23 Karuppagounder SS, Pinto JT, Xu H, Chen H-L, Beal MF, Gibson GE: Dietary supplementation with resveratrol reduces plaque pathology in a transgenic model of Alzheimer's disease. Neurochem Int 2009;54:111-118. https://doi.org/10.1016/j.neuint.2008.10.008 |
||||
| 24 Solberg NO, Chamberlin R, Vigil JR, Deck LM, Heidrich JE, Brown DC, Brady CI, Vander Jagt TA, Garwood M, Bisoffi M: Optical and SPION-Enhanced MR imaging shows that trans-stilbene inhibitors of NF-κB concomitantly lower alzheimer's disease plaque formation and microglial activation in AβPP/PS-1 transgenic mouse brain. J Alzheimers Dis 2014;40:191-212. https://doi.org/10.3233/JAD-131031 |
||||
| 25 Vergara D, Gaballo A, Signorile A, Ferretta A, Tanzarella P, Pacelli C, Di Paola M, Cocco T, Maffia M: Resveratrol Modulation of Protein Expression in parkin-Mutant Human Skin Fibroblasts: A Proteomic Approach. Oxid Med Cell Longev 2017;2017:2198243. https://doi.org/10.1155/2017/2198243 |
||||
| 26 Weterman MA, Sorrentino V, Kasher PR, Jakobs ME, van Engelen BG, Fluiter K, de Wissel MB, Sizarov A, Nürnberg G, Nürnberg P: A frameshift mutation in LRSAM1 is responsible for a dominant hereditary polyneuropathy. Hum Mol Genet 2012;21:358-370. https://doi.org/10.1093/hmg/ddr471 |
||||
| 27 Chhangani D, Upadhyay A, Amanullah A, Joshi V, Mishra A: Ubiquitin ligase ITCH recruitment suppresses the aggregation and cellular toxicity of cytoplasmic misfolded proteins. Sci Rep 2014;4:5077. https://doi.org/10.1038/srep05077 |
||||
| 28 Park D, Jeong H, Lee MN, Koh A, Kwon O, Yang YR, Noh J, Suh PG, Park H, Ryu SH: Resveratrol induces autophagy by directly inhibiting mTOR through ATP competition. Sci Rep 2016;6:1-11. https://doi.org/10.1038/srep21772 |
||||
| 29 Sato A, Okada M, Shibuya K, Watanabe E, Seino S, Suzuki K, Narita Y, Shibui S, Kayama T, Kitanaka C: Resveratrol promotes proteasome-dependent degradation of Nanog via p53 activation and induces differentiation of glioma stem cells. Stem Cell Res 2013;11:601-610. https://doi.org/10.1016/j.scr.2013.04.004 |
||||
| 30 Hillert EK, Brnjic S, Zhang X, Mazurkiewicz M, Saei AA, Mofers A, Selvaraju K, Zubarev R, Linder S, D'Arcy P: Proteasome inhibitor b-AP15 induces enhanced proteotoxicity by inhibiting cytoprotective aggresome formation. Cancer Lett 2019;448:70-83. https://doi.org/10.1016/j.canlet.2019.02.003 |
||||
| 31 Bukau B, Weissman J, Horwich A: Molecular chaperones and protein quality control. Cell 2006;125:443-451. https://doi.org/10.1016/j.cell.2006.04.014 |
||||
| 32 Ho Zhi Guang M, Kavanagh EL, Dunne LP, Dowling P, Zhang L, Lindsay S, Bazou D, Goh CY, Hanley C, Bianchi G: Targeting proteotoxic stress in cancer: a review of the role that protein quality control pathways play in oncogenesis. Cancers (Basel) 2019;11:66. https://doi.org/10.3390/cancers11010066 |
||||
| 33 Verheijen BM, Oyanagi K, Van Leeuwen FW: Dysfunction of protein quality control in parkinsonism-dementia complex of Guam. Front Neurol 2018;9:173. https://doi.org/10.3389/fneur.2018.00173 |
||||
| 34 Fu HY, Mukai M, Awata N, Sakata Y, Hori M, Minamino T: Protein quality control dysfunction in cardiovascular complications induced by anti-cancer drugs. Cardiovasc Drugs Ther 2017;31:109-117. https://doi.org/10.1007/s10557-016-6709-7 |
||||
| 35 Joshi V, Mishra R, Upadhyay A, Amanullah A, Poluri KM, Singh S, Kumar A, Mishra A: Polyphenolic flavonoid (Myricetin) upregulated proteasomal degradation mechanisms: eliminates neurodegenerative proteins aggregation. J Cell Physiol 2019;234:20900-20914. https://doi.org/10.1002/jcp.28695 |
||||
| 36 Badhwar R, Upadhyay A: Modulation of Cellular Protein Quality Control Pathways Using Small Natural Molecules; in Singh J, Meshram V, Gupta M (eds): Bioactive Natural products in Drug Discovery, Springer, 2020, pp 609-641. | ||||
| 37 Upadhyay A: Natural compounds in the regulation of proteostatic pathways: An invincible artillery against stress, ageing, and diseases. Acta Pharm Sin B 2021;11:2995-3014. https://doi.org/10.1016/j.apsb.2021.01.006 |
||||
| 38 Mishra R, Upadhyay A, Prajapati VK, Dhiman R, Poluri KM, Jana NR, Mishra A: LRSAM1 E3 ubiquitin ligase: molecular neurobiological perspectives linked with brain diseases. Cell Mol Life Sci 2019;76:2093-2110. https://doi.org/10.1007/s00018-019-03055-y |
||||
| 39 Dal Vechio FH, Cerqueira F, Augusto O, Lopes R, Demasi M: Peptides that activate the 20S proteasome by gate opening increased oxidized protein removal and reduced protein aggregation. Free Radic Biol Med 2014;67:304-313. https://doi.org/10.1016/j.freeradbiomed.2013.11.017 |
||||
| 40 Jones CL, Tepe JJ: Proteasome activation to combat proteotoxicity. Molecules 2019;24:2841. https://doi.org/10.3390/molecules24152841 |
||||
| 41 Arslan MA, Chikina M, Csermely P, Soti C: Misfolded proteins inhibit proliferation and promote stress-induced death in SV40-transformed mammalian cells. FASEB J 2012;26:766-777. https://doi.org/10.1096/fj.11-186197 |
||||
| 42 Ye M, Wu H, Li S: Resveratrol alleviates oxygen/glucose deprivation/reoxygenation‑induced neuronal damage through induction of mitophagy. Mol Med Rep 2021;23:73. https://doi.org/10.3892/mmr.2020.11711 |
||||
| 43 Higuchi-Sanabria R, Frankino PA, Paul JW, Tronnes SU, Dillin A: A Futile Battle? Protein Quality Control and the Stress of Aging. Dev Cell 2018;44:139-163. https://doi.org/10.1016/j.devcel.2017.12.020 |
||||
| 44 Mallucci G, Dickinson A, Linehan J, Klohn PC, Brandner S, Collinge J: Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science 2003;302:871-874. https://doi.org/10.1126/science.1090187 |
||||
| 45 Zheng Q, Li J, Wang X: Interplay between the ubiquitin-proteasome system and autophagy in proteinopathies. Int J Physiol Pathophysiol Pharmacol 2009;1:127-142. | ||||