De Novo Cloning and Functional Characterization of a Mechanosensitive Piezo-Like Ion Channel in the Crayfish
bDepartment of Medical Genetics, Faculty of Medicine, Hacettepe University, Ankara, Turkey
Keywords
Abstract
Background/Aims:
Mechanosensitive ion channels are the principal elements in the transduction of mechanical force to neural activity. To date, considerably fewer studies have been published about the molecular and structural properties of mechanosensitive channels. Piezo channels are the only ion channel family in eukaryotes which is selectively gated by the membrane tension. Piezo channels have been described in mammals and some other eukaryotes. However, not much information is available for the crustaceans.Materials:
Conventional cloning methods were used to clone the putative PIEZO channel mRNA in crayfish ganglia samples. HEK293T cells were transfected by the plasmid of the cloned gene for functional studies. The CDS of the mRNA translated into the protein sequence and three-dimensional structure of the channel has been calculated.Results:
An mRNA, 9378 bp, was firstly cloned from crayfish which codes a 2674 residues protein. The cloned sequence is similar to the piezo channel mRNAs reported in the other species. The sequence of the coded protein has been analyzed, and some functional domains have been identified. A three-dimensional structure of the coded protein was successfully calculated in reference to mouse piezo 1 channel protein data. A plasmid with a fluorescent protein indicator was synthesized for heterologous expression in HEK293T cells. The evoked calcium response to mechanical stimulation was not different from those observed in the control cells. However, the transfected cells were more sensitive to the gating modifier YODA-1.Conclusion:
Based on the apparent similarity in sequence, structure and functional properties to other known piezo channels, it has been proposed that cloned mRNA may code a piezo-like ion channel in crayfish.Introduction
In sensory receptors, ion channels gated specifically by a certain form of physical stimulus mediate the transduction of the stimulus into a form of neural activity. In an evolutionary sense, these proteins represent the most fundamental event in the genesis of sensation and perception. Mechanosensation might have evolved as the earliest senses, as even some unicellular organisms can display a primitive form of behavior to a mechanical stimulus [1]. In mechanoreceptor cells, it has been well documented by electrophysiology experiments that mechanical stimulation evokes changes in the membrane conductance leading to a distinct mechanosensitive ionic current [2-11]. Thus, the electrophysiology data has already indicated that a distinct group of ion channels gated specifically by a form of mechanical energy are responsible for mechano-electrical transduction. However, a candidate channel must satisfy several experimental conditions to qualify as a mechano-transducer channel [12]. The limits imposed by the genetic expression of clone genes in control cell lines present a challenge in confirming specific genes as coding for mechanotransducer channels. For example, the heterologous expression of an ion channel alpha peptide may not be sufficient to enable a mechanosensitive current if a complex with other proteins is needed to generate mechanosensitivity. Furthermore, even the generation of small and calibrated mechanical stimulus at the single cell level is itself a substantial experimental challenge [13].
Based mainly on functional studies, only a few eukaryotic ion channels or families have been proposed to be mechanosensitive [14, 15]. Among those candidate channels PIEZO family, described in the previous decade [16], is the only group that was able to fulfill the criteria to be qualified as a mechano-transducer channel [12]. In addition to numerous functional studies, the three-dimensional structure of the channels has been explored [17, 18], and several parts with functional importance have been defined [19]. The PIEZO channels have been identified in many eukaryote species. However, the presence of a PIEZO channel is yet to be confirmed in crayfish, one of the first experimental model animals for mechano-electrical transduction [2-3, 20]. The present work presents the results of a de novo cloning study of a novel PIEZO-like channel gene in the crayfish and the expression of the coded protein in the HEK293T cell lines.
Materials and Methods
Construction of cDNA library: Total RNA content of the freshly dissected abdominal muscle samples was extracted by using the conventional phenol-chloroform method. The concentration of the RNA extract was about 1.5 µg/µl as measured by a fluorometric device (Qubit, Thermo Fisher Scientific, Massachusetts, USA). RNA samples were reverse transcribed into cDNA copies by using a reverse transcriptase in the presence of random and oligo dT primers. Synthesized cDNA was ligated by using a high-efficiency ligation mix. The ligated cDNA was amplified by using SensiPhi DNA Polymerase in an isothermal reaction for two hours (Repli-g WTA kit, Qiagen, Hilden, Germany). The final concentration of the cDNA library was 0.3-0.5 µg/µl [22].
Transcriptome assembly: cDNA libraries were converted into labeled short reads of 150 bp. 100 M of the short reads was sequenced for each sample in an Illumina platform (CA, USA). Short read sequences were de novo assembled into a transcriptome assembly in the DNASTAR (Madison, USA) environment. Assembled transcripts were annotated in reference to invertebrate.136.rna.fna library .
PIEZO mRNA sequences from related species were aligned and conserved regions were defined. The presence of the conserved sequences was searched among the de novo assembled transcript draft. A set of primers was designed (Table 1). The gene fragments were amplified in PCR by using the primers and cDNA library template. Amplified fragments were sequenced for confirmation of the identity and planning of the down-streaming interventions.

Table 1: List of Primers
Rapid amplification of cDNA ends polymerase chain reaction (RACE): Gene fragments were selected for each end. Primers, designed according to the target fragments, were used in a PCR experiment to confirm the presence of the target sequence. The product of the reaction was Sanger sequenced for confirmation. By using the sequence information, a pair of nesting primers were produced for each end (Table 1). A commercially available kit (SMARTer 5’/3’ kit, Clontech, Mountain View, CA, USA) was used to reveal missing parts flanking the transcripts to the 5’ and 3’ ends (NCBI# OK513276).
Sequence analysis: PCR products were run on agarose gel electrophoresis for evaluation, and selected samples with the expected size were purified by using a purification kit (QIAquick, Qiagen, Hilden, Germany). Purified products were sequenced either in Sanger sequencing or an NGS platform Miseq (Illumina, CA, USA).
Synthesis of gene plasmids: In the present work, a plasmid was synthesized by using mScarlet backbone (Addgene plasmid # 162278) for heterologous expression studies. The original plasmid, a gift from Dr. Alfred L. George Jr. from University of Feinberg School of Medicine Chicago USA [21], was modified to house cloned mRNA instead of SCN1A. The huge size of the cloned mRNA, 9378 bp, was the major challenge hindering the successful ligation of the insert to the plasmid backbone by using conventional ligation methods [22]. A set of primers (Table 1) was designed for the Gibson cloning method [23]. A circular plasmid was constructed by using a commercially available kit (Gibson Assembly cloning kit, NEB, Massachusetts, USA). 2 ml of ligation product was mixed with 50 ml 5-alpha Competent Cells (NEB, Massachusetts, USA) and kept on ice for 30 minutes. The mixture was heat shocked at 42 °C for 30 seconds and transferred to ice for another 2 minutes. 950 ml pre-warmed SOC outgrowth medium was added, and the samples were shaken at 250 rpm, 37 °C for 1 hour. Different dilutions of the outgrowth were spread on plates and incubated at 37 °C overnight. Colonies formed on the agar were selected and inoculated to 2 ml of liquid medium containing 50 mg/ml kanamycin and grown by shaking at 250 rpm at 37 °C overnight. A control PCR was conducted on the colony medium to confirm the presence of the insert. Plasmid DNA was isolated with Nucleospin Plasmid Mini Prep Kit (Macherey—Nagel #740499, Takara Bio, Kasatsu, Japan) from liquid growth of the control positive colony. Another plasmid mPiezo1-IRES-eGFP, a gift from Ardem Patapoutian from California Institute of Technology, USA (Addgene plasmid # 80925), was used for control experiments [16].
Heterologous expression experiments: Plasmids were loaded into HEK 293T cells by using a commercial transfection kit (Lipofectamine 3000, Thermo Fisher, Massachusetts, USA). Cells that were grown in 35 mm petri dishes at a 60-70 % confluence, were exposed to 2.5 µg of the plasmid for 48 hours in an incubator at 5 % CO2 and 37 oC. Dishes were examined daily under a fluorescent microscope to detect the expression of the fluorescent protein, and the medium was refreshed.
Cells were loaded with a calcium indicator dye by incubating in 1.25 µM Fluo-4 AM solution at room temperature for 20 minutes [24] and positioned under a confocal laser scanning microscope. mScarlet positive cells were considered to express the cloned piezo channel. A microelectrode with a blunt tip was positioned just above a cell (or cell group). Mechanical stimulus was applied by gently indenting the cell with the electrode by using a micromanipulator, and emitted calcium specific signal was acquired in a continuous time series imaging mode at a rate of 4 frames per second. Experiments were conducted under various experimental conditions. Control experiments were done on mScarlet negative cells.
Calculation of channel structure: Open reading frame of the cloned gene was translated into a protein sequence. The three-dimensional structure of the protein was calculated in reference to the most similar channel structure information available in protein databases (i.e., mouse P1 channel). The efficiency of the fit was evaluated by calculating a set of scores and successful alignment of the functional domains to the estimated locations [25, 17, 26-27]. However, the similarity of the cloned sequence to the reference was not the largest one as compared to other sequences (cf. Fig. 2). Alternatively, the cloned sequence was submitted into alpha-fold platform for calculation of the three-dimensional structure of the trimeric channel with no reference [28]. The size of the cloned protein sequence was a limitation in both types of calculations and the sequence had to be segmented into smaller fragments. Alternatively, the cloned protein sequence was submitted into Swiss-Model platform (Biozentrum, University of Basel, Switzerland), which can accommodate the whole sequence for structural calculation in reference to mouse P1 channel. Once the three-dimensional structure was constructed the channel pore was calculated by using the program HOLE [29].
Results
PCR experiments were conducted on cDNA templates by using various combinations of the designed primers. Two different gene fragments, at about 300 bp in length, were amplified when F2, R2 and F3, R3 primers were combined (Table 1). BLAST analysis of the sequences indicated a similarity to those of PIEZO mRNAs reported in other species. Several PCR experiments were conducted to amplify the missing part between the fragments. Cloned parts were assembled, and those were used to design nested primers for RACE experiments in both directions. First, a 3’RACE and then a 5’RACE experiment was conducted to reveal the parts flanking the cloned transcripts to both 5’ and 3’ ends. However, the part of the initially cloned mRNA fragment was rather distant to 5’end. Several attempts had to be conducted for a successful 5’RACE experiment. Finally, a complete mRNA sequence, 9378 bp in length, was cloned. The cloned mRNA sequence was registered to the NCBI database (OK513276).
The quality of the cloned mRNA sequence was controlled for by various tests. It was observed that large repeat sequences were absent in the sequence. An amplification product of the full-length mRNA (Fig. 1A) was converted into labeled short reads and sequenced in an Illumina platform (Miseq). Shorts read sequences aligned completely along the mRNA sequence without any gaps at high coverage (Fig. 1A). Furthermore, scaffolds, aligning well to the reference sequence, were obtained when those short reads were de novo assembled. Finally, the cloned mRNA sequence was aligned to the genome draft of Cherax quadricarinatus , a related species (NCBI#: GCA_009761615.1) [30]. The sequence of the cloned mRNA had a distinct alignment score [31] to one of the contigs (VSFE01006198.1) in the genome draft. The contig was very long, 195231 bp, as compared to the size of the cloned mRNA. By using an algorithm developed in our laboratory [22], possible exons along the contig were estimated (Fig. 1C).
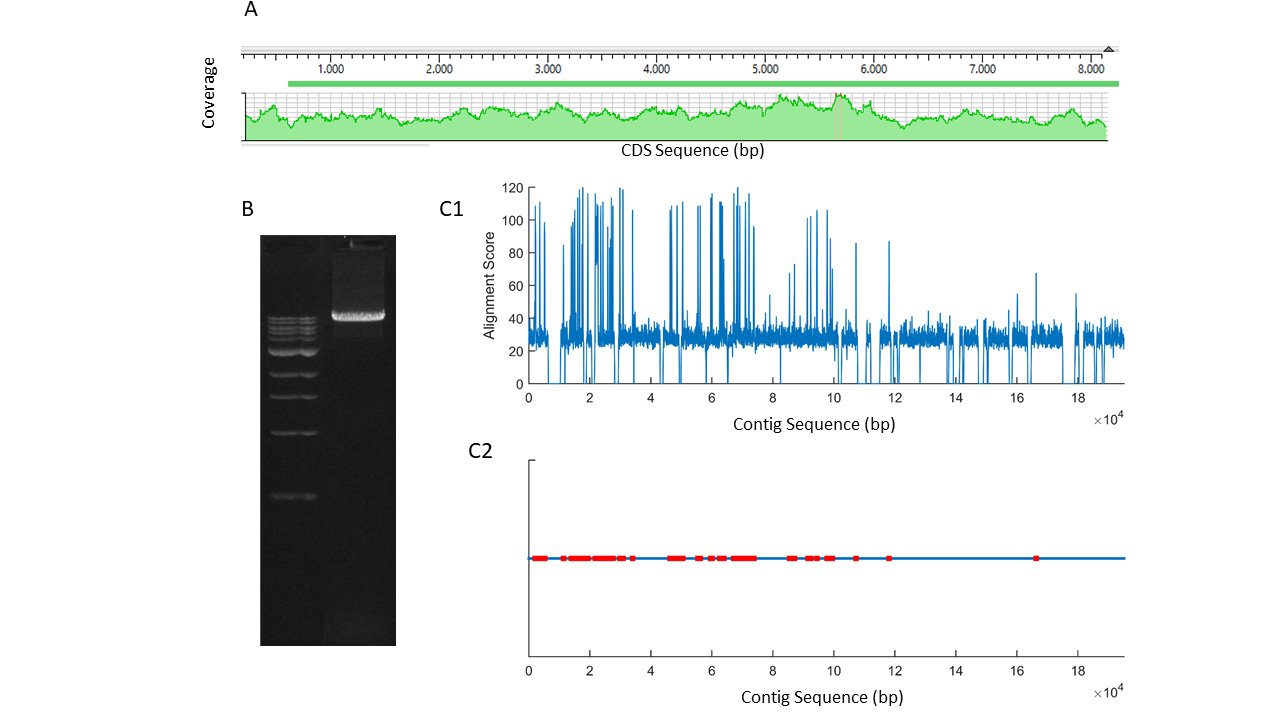
Fig. 1: A, short reads alignment score of the PCR product shown in B, obtained by using F1-R4 primers in 1/100 ganglia cDNA template (ladder 1 kb). C1, alignment scores for mers of 100 bp from a contig (VSFE01006198.1) of Cherax quadricarinatus genome draft to the cloned piezo channel mRNA. C2, localization of putative exons (red) on the contig.
The open reading frame of the cloned mRNA was located between 634-8656 residues and coded a protein with 2674 amino acids (Table 2). The protein sequence had a strong similarity to those reported for other species when compared in the Blast platform (cf. Fig. 2). It should be emphasized that the protein sequence had a better score than that obtained for the mRNA sequence. The amino acid sequence of the coded protein was compared to those in a collection of sequences from different species. 30 of those are selected from the NCBI Orthologs collections for PIEZO 1 and 2 channels, and others are the invertebrate sequences selected from the list under “similar genes” tab. A phylogenetic tree was constructed in MATLAB environment where distances were calculated by using Jukes-Cantor [32] method (Fig. 2). It was observed that a principal branching was present between animal and plant sequences. PIEZO 1, PIEZO 2 and PIEZO-like channels are grouped together in the phylogenetic tree. The cloned sequence grouped together with other invertebrate piezo-like channel sequences.
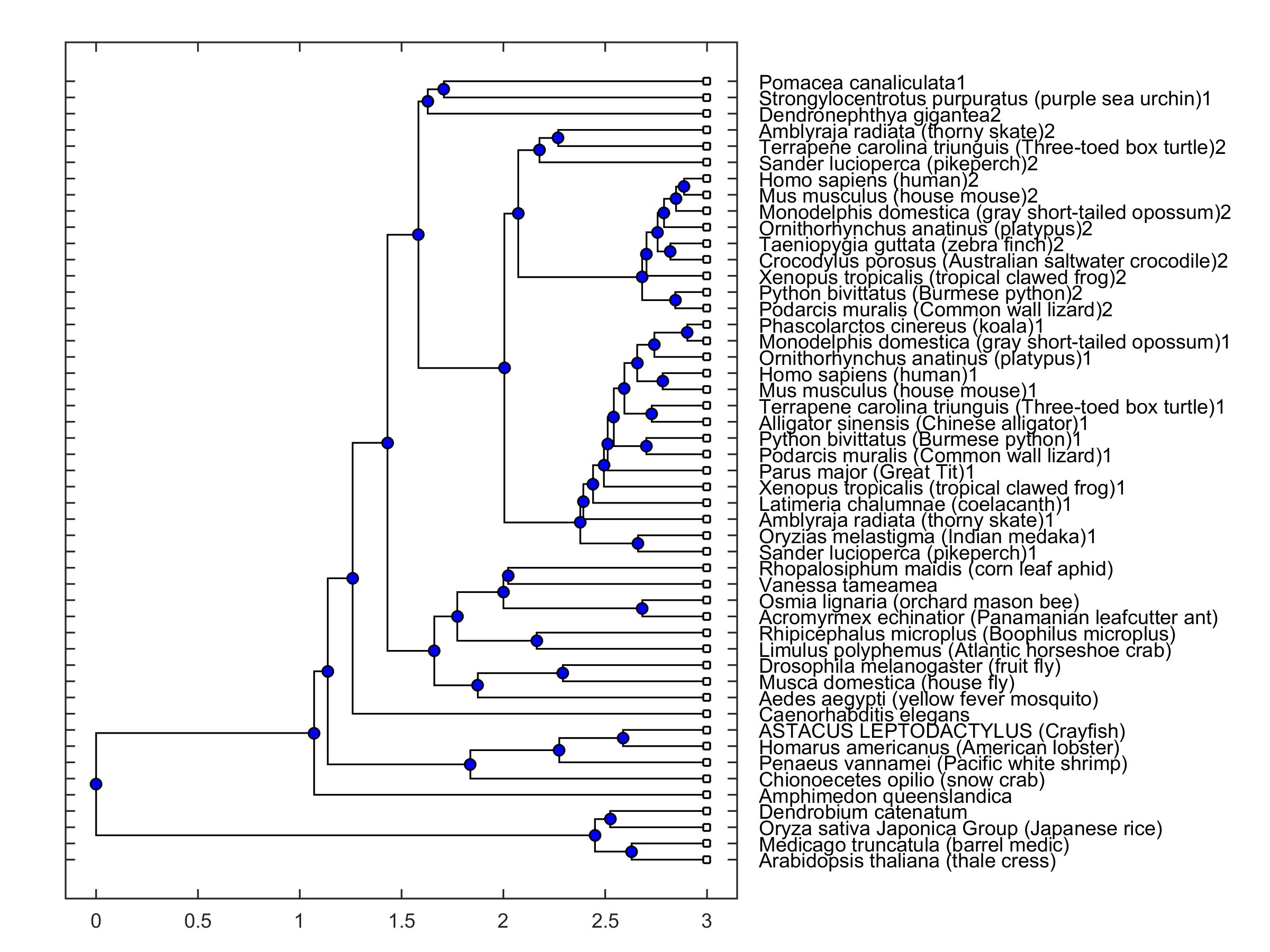
Fig. 2: Phylogenetic tree of a collection of sequences from 49 different species. Presence of 1 and 2 in labels indicates piezo1 and piezo2 type channels, respectively. Absence of 1 and 2 indicates a piezo-like channel type.
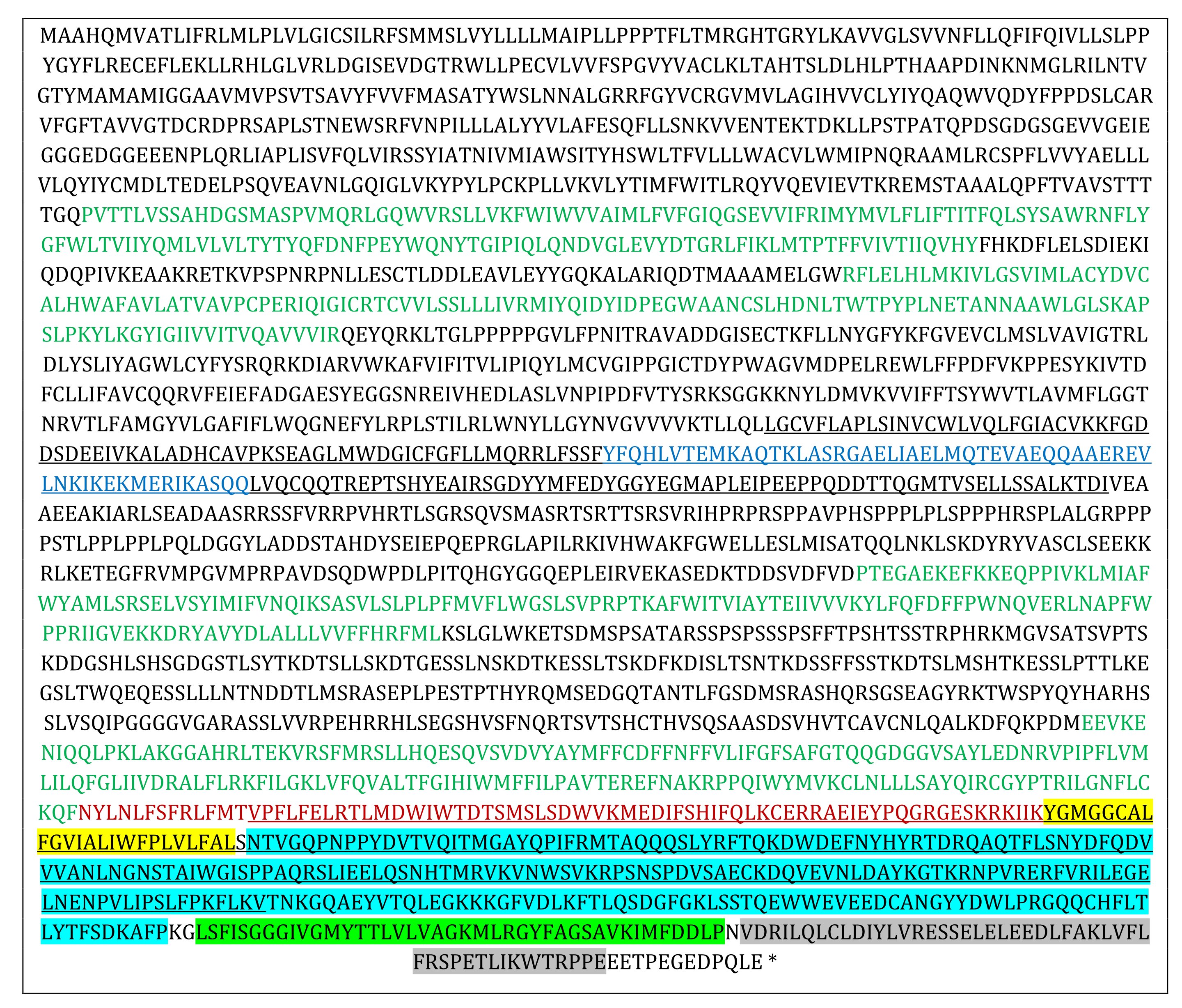
Table 2: Amino acid sequence of the coded protein. Pore module (2295-2674) is in bold. Regions 1170-1365 and 2241-2659 aligning to pfam 15917 and pfam 12166 are underlined. Regions aligning to OH, CED, IH and CTD regions defined in mouse P1 channel are highlighted in yellow, blue, green and gray, respectively. Regions aligning to beam segment of mouse P1 channel is in blue. Regions aligning to mouse P1 channel TMU 4, 5, 8, 9 are in green and “anchor” segment is in red
Functional parts of the cloned channel protein were identified in reference to those reported in the mouse P1 channel and illustrated in Table 2. By using another algorithm in the MATLAB environment, the cloned protein sequence was converted into a continuous set of kernels with a size of 20 amino acids each. Each one of the kernels was aligned [31] to mouse PIEZO 1 (mP1) and mouse PIEZO 2 (mP2) sequences (NP_001344278.1; NP_001034574.4) which is one of the most extensively studied PIEZO sequences. Alignment scores are given in Fig. 3. The cloned protein sequence was subjected to a hydrophobicity test [33] as the window length and edge weight were 20 and 0.95, respectively (Fig. 3B). It was observed that the similarity score of the cloned channel protein to mouse P1 and P2 channels appeared to be high for the hydrophobic parts as compared to the hydrophilic parts. The correlation coefficient of the comparison between the similarity score to P1 and the hydrophobicity score for the whole sequence was 0.39. However, the correlation substantially increased to 0.58 when the analysis was restricted to the pore module only.
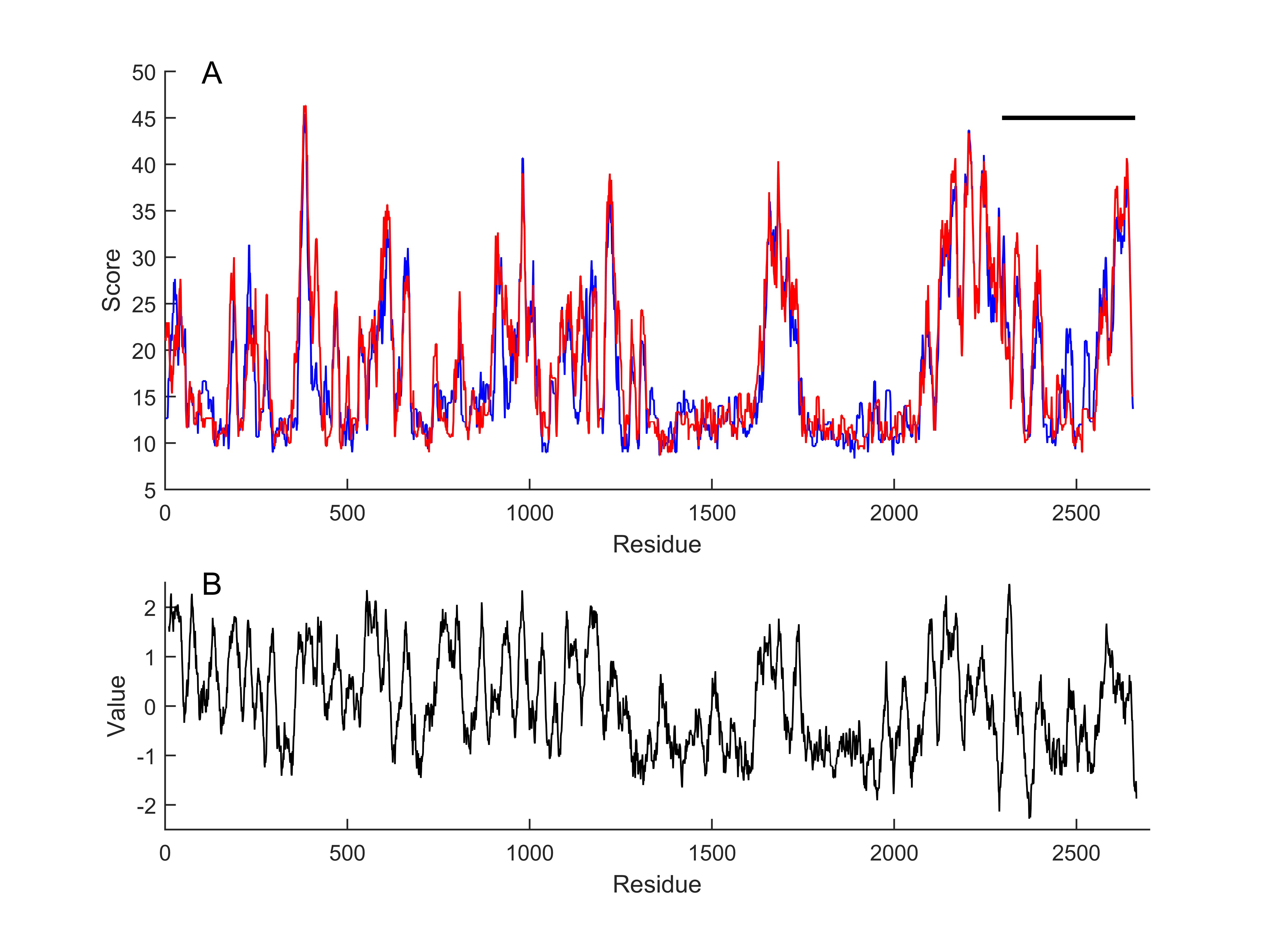
Fig. 3: Local alignment scores of continuous set of kernels, with a size of 20 amino acids, to mouse piezo1 (blue) and piezo2 (red) sequences, respectively (A). Horizontal line indicates putative pore module of the cloned channel. Kyte-Doolittle hydrophobicity score of the cloned channel (B). Window length 20, linear filter at edge width of 0.95.
Based on the primary amino acid sequence three-dimensional protein structure of the monomer and trimeric channel were calculated. I-TASSER (University of Michigen, Medical School, USA) and Alphafold Colab (DeepMind Technologies, London, UK) platforms do not accommodate the large protein sequence given in Table 2. The cloned channel sequence was split into two pieces so that the beam segment was common. Each one of them was independently submitted into the I-TASSER platform with reference to the mouse P1 channel structure. Calculated models were aligned onto each other along the beam segment and assembled to get the structure of the whole monomer (Figure S1). Alternatively, the pore region of the cloned protein sequence was submitted into the Alphafold Colab platform to obtain a partial calculation of the trimeric channel structure (Figure S1). However, both facilities gave some incomplete calculations. Swiss-Model platform was capable of calculating the whole trimeric channel in a single session with reference to the mouse P1 channel data (5z10.pdb) (Fig. 4A and B). The calculated 3D structure of the cloned channel was very similar to the reference (Fig. 4C). Similarity and coverage of the sequence to the reference (5z10.pdb) were 0.40 and 0.49, respectively. QSQE score of the calculation was 0.54. However, the similarity, coverage, and QSQE scores were 0.41, 0.93, and 0.91 when only the putative pore module was submitted with the same reference. The pore of the channel was calculated in the model as F residues at the 2573rd position forming the narrowest part (Fig. 4D).
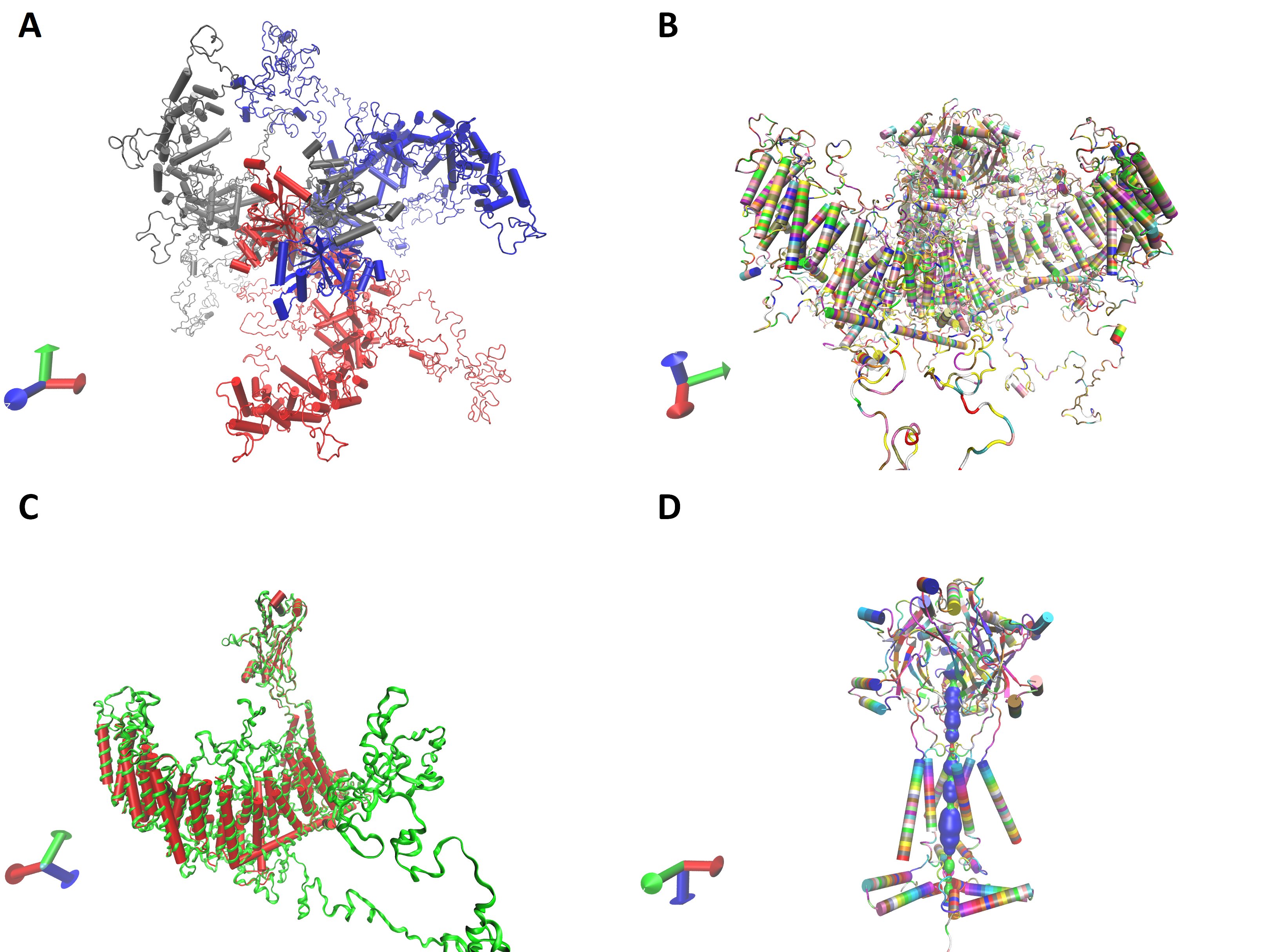
Fig. 4: Calculated three dimensional structure of the cloned channel protein. A and B, trimeric model from top and lateral view, in chain and residue name mode, respectively. C, is monomeric structure of the channel protein (green) as superimposed on the reference, mouse P1, channel protein structure (red). Trimeric model of the pore module with the calculated pore (D).
The cloned PIEZO-mScarlet plasmid was expressed in HEK293T cells (Fig. 5). The presence of a red fluorescent signal was interpreted as a successful expression of the cloned channel protein (Fig. 5B). Cells were loaded with calcium indicator Fluo-4-AM by incubation (Fig. 5A). Mechanical stimulation by gently poking the cells with a blunt electrode evoked a rise in calcium sensitive emission signal (Fig. 5C). The rise of the signal was rapid, while the decay was considerably slower and incomplete (Fig. 5D). Repetitive stimulation of the same cell evoked repetitive calcium transients if sufficiently long resting periods were allowed in between the stimulations. Mechanical stimulation of the PIEZO-mScarlet negative and positive cells evoked a rapid rise in calcium specific Fluo-4 emission signal (Fig. 5E). Variance in amplitude and the waveform was large. The recorded signals were normalized to compare timing calcium signals to mechanical stimuli (Fig. 5F). Duration of the rise was 9.63 ± 1.3 s (Mean ± SEM, n=12) and 7.84 ± 1.02 (Mean ± SEM, n=12) in the mScarlet positive and negative cells, respectively. However, a statistically significant difference was not observed when responses recorded in PIEZO-mScarlet positive cells were compared to those from the negative cells (Fig. 5F). The PIEZO-mScarlet expressing cells were exposed to P1 channel gating modifier YODA-1 together with the control cells. Exposure to YODA-1 evoked a rise in resting calcium levels in both transfected and control cells. The rise was significantly larger in transfected cells as compared to the control cells (Fig. 5F). Further, in some of the transfected cells, calcium transients were observed in addition to the slowly developing rise in response to the YODA-1 exposure.
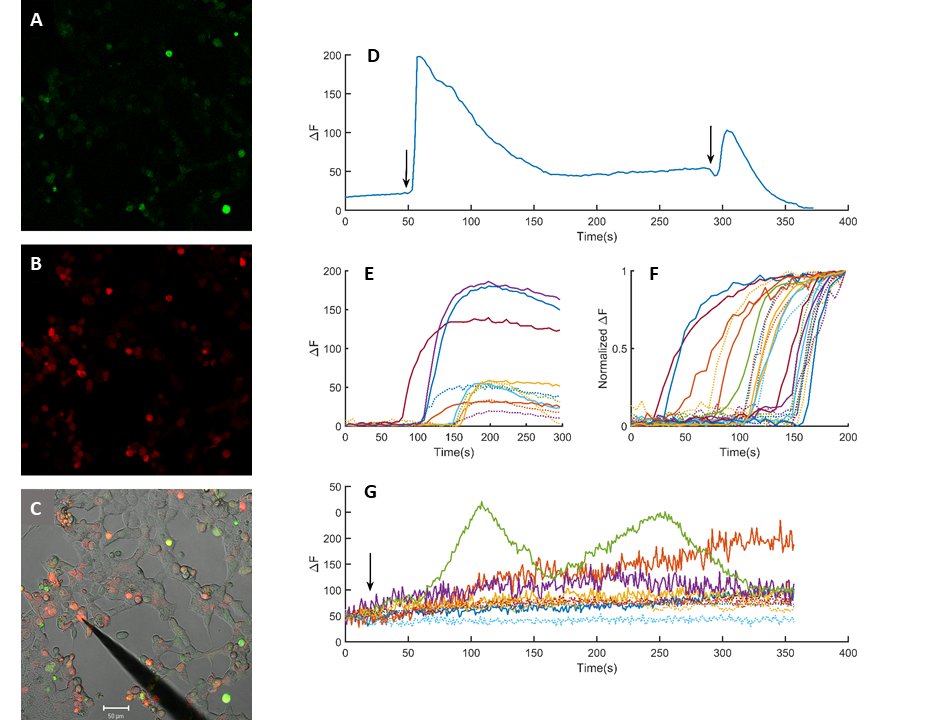
Fig. 5: Heterologous expression of the cloned piezo channel. HEK293T cells expressing mScarlet-piezo vector (B) after incubation in Fluo4-AM (1.25 µM) for 30 minutes (A). C is the superimposed picture of A, B and a transmitted light image of the same window. Scale bar is 50 µm. Intracellular calcium transients in response to mechanical stimuli (D). Comparison of a sample base line corrected calcium transients to mechanical stimulation in control and mScarlet-piezo expressing cells (E) dotted and solid lines, respectively.(F) Normalized calcium transients in control (n=12) and mScarlet expressing cells (n=12) dotted and solid lines, respectively. Changes in calcium levels in response to 70 µM YODA-1 exposure (G). Arrows indicate application of mechanical stimuli in D and YODA-1 exposure in F.
Cloned channel protein expressed in HEK 293T cells were not labeled by a mouse p1 specific polyclonal antibody (15939-1-AP, Proteintech), while mouse P1 channel protein expressing cells were specifically labeled (Figure S2).
Discussion
The result of cloning experiments has revealed an original mRNA sequence from the cDNA library from Astacus leptodactylus (Table S1). The length of the sequence is 9378 bp, and the ORF, located between 634-8656 residues, coded a protein with 2674 amino acids (Table 2). Cloned mRNA was subjected to various quality checks (Fig. 1). Sequence analysis of the cloned mRNA and calculated protein sequence indicated a substantial similarity to those in PIEZO type ion channels explored in other species ranging from invertebrates to mammals and plants (Fig. 2 and 3). The calculated protein sequence possesses two conserved domains, pfam15917 and pfam12166, which are related to PIEZO channel proteins [34]. Those domains align between 2241- 2659 and 1170-1365 residues in the cloned channel protein, respectively (Table 2). Thus, it is conceivable that the cloned mRNA sequence might be related to a PIEZO type ion channel present in our model animal. The mouse mechanosensitive PIEZO 1 and 2 channels are both transmembrane proteins with a highly complex structure different from all other ion channels [17, 35-36]. PIEZO channel has a three bladed, propeller like homotrimeric architecture with a pore domain in its center. A substantial amount of information about functional parts of the channel has become available through some combinations of molecular biology interventions and high-resolution structural methods in recent years [18, 19, 35, 37-39]. Thus, some functional parts of the putative channel protein have been estimated in considering those studies and focusing on the structure-function relationship in mP1 channels. Mouse P1 channel pore module (2189-2547), consists of an outer helix (OH, 2189-2213), a c terminal extracellular domain (CED, 2214-2457), an inner helix (2458-2499) and and intracellular c terminal domain (CTD, 2500- 2547) segments [17, 19, 39]. A segment of the cloned sequence, between the residues 2302-2662, aligned well to the pore module of the mP1 channel (Table 2). Further, the alignment region included the sequence of a conserved domain, pfam12166, between the residues 2241-2659. As shown in Table 2, the identified functional part of the mouse piezo channel aligned well with the distinct parts of the cloned sequence. The sequence of the IH segment, building the inner walls of the pore in the mP1 channel, “ivglyvsivlvvgkfvrgffseishsimfeelpc” aligns to the “IVGMYTTLVLVAGKMLRGYFAGSAVKIMFDDLP” sequence in the cloned sequence. The presence of strong polar residues like DD, K, and LV in the segment indicates that IH might be related to voltage dependent inactivation as in the mP1 channel. In the CED segment of the cloned sequence is absent a “DEEED” motif which is related to ion selectivity in the mP1 channel. Instead a ”EVEED“ motif is present. However, the location of the polar residues is different from that in the mP1 channel.
Many “E” and “D” residues are present in the estimated CTD region. It is not possible to assign a functional property to those residues at present. However, in mP1 channel mutations of those residues alter ion selectivity and ruthenium red sensitivity [19]. “IMFDDLP” and “NVD” residues may have a similar function.
The outer helix of the mP1 channel is related to the mechanosensitive property [39]. Some of the parts of the domain align well with the cloned channel sequence. The beam segment of the mP1 channel, which is related to mechanotransduction and Yoda and Jedi1/2 sensitivity, aligns the segment between 1243-1299 residues of the cloned sequence. However, mechanosensation is also related to the transmembrane unit (TMU) 4 and 5 in the mP1 channel [19, 38-39]. Those regions align to segments between 517-671 and 746-879 residues, respectively. Similarly, TMU8 and TMU9 related Yoda sensitivity aligns to segments between 1603-1742 and 2047-2227 residues, respectively. However, alignments in the cloned sequence have a gap in between (cf. Table 2) while they are continuous in the mP1 sequence. Similarly, the “anchor” sequence of the mP1 channel, related to ion selection and ruthenium red sensitivity aligns to residues between 2128-2301 in the cloned sequence.
The sequence similarity of the cloned protein to mP1 and mP2 channels is similar (Fig. 3). The most similar kernel “LLSAYQIRCGYPTRILGNFL” aligns almost completely with the reference sequences out of the initial L residue. This segment overlaps with the putative THU9 segment related to the Yoda sensitivity in the mP1 channel. Comparison to mouse piezo channel is a rational reference to estimate possible functions of the cloned channel, such as being responsive to mechanical stimulus or possessing a sensitivity to RR and Yoda exposure. Further, the relevancy of the estimate was experimentally confirmed by the observation that the transfected cells are Yoda sensitive (Fig. 5). Based on the similarity, a three-dimensional structure of the channel could be calculated with reference to the mouse P1 channel (Fig. 4). It was observed that the transmembrane segments of the cloned channel fit into the reference better than the other parts (Fig. 4C). The finding is supportive of the 2D analysis data indicating that the hydrophobic regions, likely to be the transmembrane segments, align better to mouse piezo channels (Fig. 3). Thus, the amino acid sequence of the transmembrane segments of the piezo channels might have been conserved at a larger rate as compared to the other parts of the channel (Table 2 and Fig. 3). It should be pointed out that the cloned channel protein is 127 residues larger in length than the reference mouse P1 channel. Size discrepancy is another weakness leading to relatively poor scores, particularly in the three-dimensional calculations (Fig. 4). The pore module of the channel is a highly conserved region among species [17, 40]. A significant similarity to even other types of trimeric channels has also been reported [41-43]. We obtained a very high QSQE score (0.91) in three-dimensional calculations when only the pore module, having a high hydrophobicity score (cf. Fig. 2) and high coverage to the reference (0.93), was used (Fig. 4D). The cloned sequence lacks repeat motifs larger than six amino acids (Table 2). Thus, functional repeat motifs, as in the drosophila NOMPC channel, is not present in the cloned sequence [40].
Sequence analysis and the structural calculations of the cloned protein indicate that a putative piezo-like channel gene was cloned. Though calcium signals were larger and longer in the transfected cells the difference is not statistically significant (Fig. 5E). The native cells have a capacity to develop a rise in the cytosolic calcium when mechanically stimulated (Fig. 5D). Further, production of a calibrated mechanical stimulus is a very elaborate experimental intervention [13]. We estimate that the poking the cell with an electrode might simultaneously activate the native and heterelogously expressed components, both leading to a rise in the calcium concentration. Thus, the overlapping components can’t be resolved efficiently.
Conclusion
In the present work, an mRNA coding a protein with 2674 residues was originally cloned. Based on the apparent similarity in sequence, structure, and functional properties to other known piezo channels, it has been proposed that cloned mRNA may code a piezo-like ion channel in the crayfish.
Acknowledgements
Author Contributions
Authors equally contributed to the present work. The authors are grateful to Dr. Daniel Press for reading the manuscript.
Funding Sources
The research was supported by grants from Hacettepe University Research Foundation (HU project 15403, 19942) and The Scientific and Technological Research Council of Turkey (TUBITAK project 218S553). The authors thank the anonymous reviewers.
Statement of Ethics
The local Institutional Review Board deemed the study exempt from review.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
| 1 | Grell K. Protozoologie. Springer-Verlag Berlin 1956; 284 pp.
https://doi.org/10.1007/978-3-642-49860-2 |
| 2 | Alexandrowicz JS: Muscle receptor organs in the abdomen of Homarus vulgaris and Palimurus vulgaris. Quart J Microsc 1951;92:163-200.
https://doi.org/10.1242/jcs.s3-92.18.163 |
| 3 | Wiersma CAG, Furshpan E, Florey E: Physiological and pharmacological observations on muscle receptor organs of the crayfish, Cambarus clarkia Girard. J Exp Biol 1953;30:136-150.
https://doi.org/10.1242/jeb.30.1.136 |
| 4 | Hudspeth AJ, Corey DP: Sensitivity, polarity, and conductance change in the response of vertebrate hair cells to controlled mechanical stimuli. Proc Natl Acad Sci 1977;74:2407-2411.
https://doi.org/10.1073/pnas.74.6.2407 |
| 5 | Eckert R, Brehm P: Ionic mechanism of excitation in Paramecium. Annu Rev Biophys Bioeng 1979;8:353-383.
https://doi.org/10.1146/annurev.bb.08.060179.002033 |
| 6 | Hoyle G. In electrical Conduction and Behaviour in Simple Invertebrates. G. A. B. Shelton (ed.) Calearendon Press 1982;pp:1-48.
|
| 7 | Guharay F, Sachs F: Stretch activated sing ion channel currents in tissue cultered embryonic chick skeletal muscle. J Physiol 1984;352:685-701.
https://doi.org/10.1113/jphysiol.1984.sp015317 |
| 8 | Wood DC: The functional significance of evolutionary modifications found in ciliate, Stentor. In Evolution of the First Nervous Systems, P. A. V. Anderson (Ed.) NATO ASI Series A, Life Sciences 1989;188:357-371.
https://doi.org/10.1007/978-1-4899-0921-3_26 |
| 9 | Deitmer J: The functional significance of evolutionary modifications found in ciliate, Stentor. In Evolution of the First Nervous Systems, P. A. V. Anderson (Ed.) NATO ASI Series A, Life Sciences 1989;188:255-265.
|
| 10 | Erxleben C: Stretch-activated current through single ion channels in the abdominal stretch receptor organ of the crayfish. J Gen Physiol 1989;94:1071-1083.
https://doi.org/10.1085/jgp.94.6.1071 |
| 11 | Rydqvist B, Purali N: Transducer properties of the rapidly adapting stretch receptor neurone in the crayfish (Pacifastacus leniusculus). J Physiol 1993;469:193-211.
https://doi.org/10.1113/jphysiol.1993.sp019811 |
| 12 | Arnadóttir J, Chalfie M: Eukaryotic mechanosensitive channels. Annu Rev Biophys 2010;39:111-137.
https://doi.org/10.1146/annurev.biophys.37.032807.125836 |
| 13 | Sachs F: Mechanical transduction by ion channels: a cautionary tale. World J Neurol 2015;5:74-87.
https://doi.org/10.5316/wjn.v5.i3.74 |
| 14 | Ranade SS, Syeda R, Patapoutian A: Mechanically Activated Ion Channels. Nueron 2015;87:1162-1189.
https://doi.org/10.1016/j.neuron.2015.08.032 |
| 15 | Kefauver JM, Ward AB, Patapoutian A: Discoveries in structure and physiology of mechanically activated ion channels. Nature 2020;587:567-587.
https://doi.org/10.1038/s41586-020-2933-1 |
| 16 | Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE, Patapoutian A: Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 2010;330:55-60.
https://doi.org/10.1126/science.1193270 |
| 17 | Ge J, Li W, Zhao Q, Li N, Chen M, Zhi P, Li R, Gao N, Xiao B, Yang M: Architecture of the mammalian mechanosensitive Piezo 1 channel. Nature 2015;527:64-69.
https://doi.org/10.1038/nature15247 |
| 18 | Guo YSR, MacKinnon R: Structure-based membrane dome mechanism for Piezo mechanosensitivity. eLife 2017;6:e33660.
https://doi.org/10.7554/eLife.33660 |
| 19 | Zhao Q, Wu K, Geng J, Chi S, Wang Y, Zhi P, Zhang M, Xiao B: Ion Permeation and Mechanotransduction Mechanisms of Mechanosensitive Piezo Channels. Neuron 2016;89:1248-1263.
https://doi.org/10.1016/j.neuron.2016.01.046 |
| 20 | Eyzaguirre C, Kuffler SW: Processes of excitation in the dentrites and in the soma of single isolated sensory nerve cells of the lobster and crayfish. J Gen Physiol 1955;39:87-119.
https://doi.org/10.1085/jgp.39.1.87 |
| 21 | DeKeyser JM, Thompson CH, George Jr. AL: Cryptic prokaryotic promoters explain instability of recombinant neuronal sodium channels in bacteria. J Biol Chem 2021;296:100298.
https://doi.org/10.1016/j.jbc.2021.100298 |
| 22 | Ergin B, Saglam B, Taskiran ZE, Bastug T, Purali N: De novo cloning and functional characterization of potassium channel genes and proteins in the crayfish Astacus leptodactylus (Eschscholtz, 1823) (Decapoda: Astacidea:Astacidea). J Crust Biol 2022;42:1.
https://doi.org/10.1093/jcbiol/ruac018 |
| 23 | Gibson DG, Young L, Chuang RY, Craig V, Hutchison III CA, Smith HO: Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 2009;6:343-347.
https://doi.org/10.1038/nmeth.1318 |
| 24 | Kayman Kürekçi G, Kural Mangit E, Koyunlar C, Unsal S, Saglam B, Ergin B, Gizer M, Uyanik I, Boustanabadimaralan Düz N, Korkusuz P, Talim B, Purali N, Hughes SM, Dincer PR: Knockout of zebrafish desmin genes does not cause skeletal muscle degeneration but alters calcium flux. Sci Rep 2021;11:7505.
https://doi.org/10.1038/s41598-021-86974-w |
| 25 | Humprey W, Dalke A, Schulten K. VMD: visual molecular Dynamics. J Mol Graph 1996;14: 33-8,27-28.
https://doi.org/10.1016/0263-7855(96)00018-5 |
| 26 | Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y: The I-TASSER Suite: Protein structure and function prediction. Nature Methods 2015;12:7-8.
https://doi.org/10.1038/nmeth.3213 |
| 27 | Zheng W, Zhang C, Li Y, Pearce R, Bell EW, Zhang Y: Folding non-homology proteins by coupling deeplearning contact maps with I-TASSER assembly simulations. Cell Reports Methods 2021;1:100014.
https://doi.org/10.1016/j.crmeth.2021.100014 |
| 28 | Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M: ColabFold - making protein folding accessible to all. Nature Methods 2022;19:679-682.
https://doi.org/10.1038/s41592-022-01488-1 |
| 29 | Smart SO, Neduvelil JG, Wang X, Wallace BA, Sanson MSP: HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 1996;14:354-360.
https://doi.org/10.1016/S0263-7855(97)00009-X |
| 30 | Tan MH, Gan HM, Lee YP, Grandjean F, Croft LJ, Austin CM: A Giant Genome for a Giant Crayfish (Cherax quadricarinatus) With Insights Into cox1 Pseudogenes in Decapod Genomes. Frontier in Genetics. 2020;11:201.
https://doi.org/10.3389/fgene.2020.00201 |
| 31 | Smith T, Waterman M: Identification of common molecular subsequences. J Mol Biol 1981;147:195-197.
https://doi.org/10.1016/0022-2836(81)90087-5 |
| 32 | Jukes TH, Cantor CR: Evolution of Protein Molecules. New York: Academic Press 1969;21-132.
https://doi.org/10.1016/B978-1-4832-3211-9.50009-7 |
| 33 | Kyte J, Doolittle RF: A simple method for displaying the hydropathic character of a protein. J Mol Biol 1982;157:105-132.
https://doi.org/10.1016/0022-2836(82)90515-0 |
| 34 | Kamajaya A, Kaiser JT, Lee J, Reid M, Rees DC: The structure of a conserved piezo channel domain reveals a topologically distinct beta sandwich fold. Structure 2014;22:1520-1527.
https://doi.org/10.1016/j.str.2014.08.009 |
| 35 | Saotome K, Murthy S, Kefauver JM, Whitwam T, Patapoutian A, Ward AB: Structure of the mechanically activated ion channel Piezol. Nature 2018;554:481486.
https://doi.org/10.2210/pdb6bpz/pdb |
| 36 | Wang L, Zhou H, Zhang M, Liu W, Deng T, Zhao Q, Li Y, Lei J, Li X, Xiao B: Structure and mechanogating of the mammalian tactile channel PIEZO2 Nature 2019;573:225-229.
https://doi.org/10.1038/s41586-019-1505-8 |
| 37 | Xu XZ: Demystifying mechanosensitive piezo ion channels. Neurosci Bull 2016;32:307-309.
https://doi.org/10.1007/s12264-016-0033-x |
| 38 | Zhao Q, Zhou H, Li X, Xiao B: The mechanosensitive Piezo1 channel: a three-bladed propeller-like structure and a lever-like mechanogating mechanism. FEBS J 2019;286:2461-2470.
https://doi.org/10.1111/febs.14711 |
| 39 | Jiang Y, Xuzhong Y, Jiang J, Xiao B: Structural designs and mechanogating mechanisms of the mechanosensitive piezo channels. Trends in Biochemical Sciences 2021;46:472-488.
https://doi.org/10.1016/j.tibs.2021.01.008 |
| 40 | Zhang W, Cheng LE, Kittelmann M, Li J, Petkovic M, Cheng T, Jin P, Guo Z, Göpfert MC, Jan LY, Jan YN: Ankyrin Repeats Convey Force to Gate the NOMPC Mechanotransduction Channel. Cell 2015;162:1391-1403.
https://doi.org/10.1016/j.cell.2015.08.024 |
| 41 | Baconguis I, Gouaux E: Structural plasticity and dynamic selectivityof acid-sensing ion channel-spider toxin complexes. Nature 2012;489:400-405.
https://doi.org/10.1038/nature11375 |
| 42 | Li T, Yang Y, Canessa CM: Outlines of the pore in open and closed conformations describe the gatingmechanism of ASIC1. Nat Commun 2011;2:399.
https://doi.org/10.1038/ncomms1409 |
| 43 | Gonzales EB, Kawate T, Gouaux E: Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature 2009;460:599-604.
https://doi.org/10.1038/nature08218 |