Original Article - DOI:10.33594/000000668
Accepted 24 October 2023 - Published online 13 November 2023
Docosahexaenoic Acid (DHA) Reduces LPS-Induced Inflammatory Response Via ATF3 Transcription Factor and Stimulates Src/Syk Signaling-Dependent Phagocytosis in Microglia
Keywords
Abstract
Background/Aims:
Microglial cells play a crucial role in the development of neuroinflammation in response to harmful stimuli, such as infection, ischemia or injury. Their chronic activation, however, is associated with a progression of neurodegenerative diseases. Therefore, looking for potential factors limiting microglial activation, the effect of docosahexaenoic acid (DHA) on the inflammatory response and TREM2-dependent phagocytic activity in microglia was investigated.Methods:
In LPS-induced primary microglia preincubated with DHA, or without preincubation the expression of ATF3 and TREM2 genes and TREM2, Syk, Akt proteins were determined by RT-PCR and WB, respectively. Cell viability was assayed by MTT and cytokine and chemokine expression was determined by the Proteome Profiler assay. Moreover, the phagocytic activity of microglia was assayed using immunofluorescence.Results:
We found that DHA significantly increased the expression of ATF3 , and decreased the levels of CINC-1, CINC-2αβ, CINC-3 chemokines, IL-1α and IL-1β cytokines, and ICAM-1 adhesion protein. Additionally, preincubation of microglia with DHA resulted in increased Src/Syk kinases activation associated with increased phagocytic microglia activity.Conclusion:
These findings indicate that DHA efficiently inhibits ATF3-dependent release of proinflammatory mediators and enhances phagocytic activity of microglia. The study provides a new mechanism of DHA action in reactive microglia, which may help limit neuronal damage caused by the pro-inflammatory milieu in the brain.Introduction
Microglia, the macrophages of the central nervous system (CNS), are essential for protection of the brain parenchyma against inflammation and infection. During the neonatal and juvenile development of the CNS they support myelination, oligodendrogenesis, neurogenesis and axon fasciculation through the synthesis and release of cytokines and growth hormones [1–3]. Microglia can also stimulate dendritic spine and synapse formation, releasing brain-derived neurotrophic factor (BDNF), prostaglandin E2 (PGE2) and IL-10 [4–6]. In the adult brain, microglia are essential for antibacterial and antiviral defense, as well as for elimination of cell debris, redundant synapses and protein aggregates harmful for neurons [7, 8]. In response to the harmful stimuli, microglia change their morphology, increase the expression of COX2 and iNOS, and release pro-inflammatory cytokines, such as TNF-α, IL-6, IL-1β, TGF-β, which contribute to various aspects of neuroinflammation [9–11]. Undoubtedly, phagocytosis and immunomodulation are the most important functions of microglia, and their dysregulation leads to CNS malfunction and contributes to the progression of neurological diseases, such as Alzheimer’s (AD) and Parkinson’s diseases.
TREM2 (Triggering receptor expressed on myeloid cells 2) is a ∼40-kDa type I membrane glycoprotein that is believed to be exclusively expressed on microglia [12, 13]. This membrane receptor binds various ligands, such as phosphatidylserine (PS), and phosphatidylethanolamine (PE) [14], sphingosine-1-phosphate [15], low-density and high-density lipoproteins [16], Hsp 60 [17], apolipoprotein E (ApoE) [18] or amyloid β (Aβ) [19]. As a result of TREM2 activation, the adaptor proteins, DAP12 or DAP10, interact with TREM2 via their transmembrane lysine and aspartic acid, followed by tyrosine Src kinase phosphorylation of the specific motifs in the adaptor proteins, the immunoreceptor tyrosine-based activation motif (ITAM), or the tyrosine–isoleucine–asparagine–methionine motif (YINM). Phosphorylation of the adaptor protein motifs creates the docking sites for Syk and PI3K kinases [20] which triggers a downstream signaling involved in various cellular processes, proliferation, survival, phagocytosis, production of pro-inflammatory cytokine, and cytoskeletal rearrangement [21]. It has been demonstrated that mutations in TREM2 are associated with a greater risk of earlier and more rapid development of early-onset dementia and AD [22, 23]. To date, more than 40 TREM2 mutations have been identified that are associated with microglial dysfunction, in particular an impaired phagocytosis [24]. Moreover, a diminished phagocytic clearance of amyloid plaques by microglia in AD mice models with TREM2 knockout gene was demonstrated [25, 26]. Thus, a modification of TREM2 downstream signalling is of primary interest as a potential target for a new strategy of neurodegenerative disease treatment.
Cumulative evidence indicates that n-3 PUFAs, including docosahexaenoic acid (DHA), are powerful anti-inflammatory nutrients [27]. They regulate synthesis and release of pro-inflammatory mediators [28, 29] and their supplementation significantly decreases the circulating inflammatory markers and oxidative stress [30]. Numerous studies have demonstrated that DHA and its derivatives down-regulate the expression of the pro-inflammatory mediators and at the same time, up-regulate the anti-inflammatory ones [31, 32]. Consequently, a dietary n-3 PUFAs supplementation inhibited LPS-induced increase of IL-1β and TNF-α in the mouse hippocampus [33]. Moreover, it has been demonstrated that DHA and its derivative RvD1 stimulate Aβ phagocytosis by microglia [34, 35].
However, the anti-inflammatory effect and modulation of phagocytic activity of microglia by DHA via TREM2 receptor signaling is not fully understood yet. Therefore, we investigated the effect of DHA on the pro-inflammatory cytokines and TREM2 expression, and activation of the down-stream TREM2 receptor kinases, Src and Syk, involved in phagocytosis by activated microglia.
Materials and Methods
Microglia culture
Microglia were isolated from the cerebral cortex of 1-2 days old Wistar rat pups. The brain was removed from the skull, cleaned from the meninges, and brain tissue was dispersed by repeated triturating with a pipette. Cell suspension was filtered through Nitex 180 µm and 30 µm filters (Merck, Millipore), and centrifuged at 800 rpm for 5 min. Supernatant was discarded while cells were counted and seeded into polylysine-coated 75 cm2 culture flasks (3x106 cells/flask) in DMEM containing 10% FBS and 10% horse serum. After 10 days without medium changing microglia were isolated by shaking the flasks on an orbital shaker at 240 rpm for 3 h at 37°C. The supernatant was collected and centrifuged at 1000 rpm for 10 min at RT. Pellet was resuspended, cells were plated in polylysine-coated culture plates in DMEM containing 10% FBS and 10% horse serum and then used for experiments.
MTT assay
The colorimetric MTT assay based on the cleavage of yellow tetrazolium salt (MTT) to purple formazan was used. The cells were seeded in 96-well microplates at density of 3x104 cells/well for 24 h. Then they were incubated in serum-free DMEM with DHA at concentrations from 5 µM to 50 µM for 12h and LPS at concentrations from 2.5 to 1000 ng/ml for 2.5 h. Control cells were cultured in serum-free DMEM w/o DHA and LPS. Next, medium was discarded, cells washed twice, and MTT solution at final concentration 0.5 mg/ml was added. The cells were incubated in the dark for 2 h, the solution was aspirated, then the formazan crystals were dissolved in DMSO, and the absorbance was measured at 570 nm using plate reader (Victor2, Perkin-Elmer). The percentage of viable cells was calculated with respect to control cells.
Cytokines and chemokines detection
Cytokines and chemokines in microglia were detected using the Proteome Profiler Cytokine Array Panel A (R&D Systems) which recognizes cytokines, chemokines, and other soluble mediators (Table 1). The cells were cultured in 9 cm Petri dishes (11x106 cells) and incubated with 20 µM DHA, or control medium for 12 h and then activated with LPS (10 ng/ml) for 2.5 h. Next, the cells were lysed with M-PER Reagent according to manufacturer’s instruction and lysates were incubated with the array membranes containing the assayed cytokine/chemokine antibodies for 18 h at 4°C. After several washings, the membranes were incubated with secondary antibodies conjugated with horseradish peroxidase (HRP)and exposed to HRP substrate. The images were documented by the ChemiDoc MP Imager (BioRad) and analyzed densitometrically with the ImageJ software (National Institute of Health, USA) using the Dot Blot Analyzer plugin.
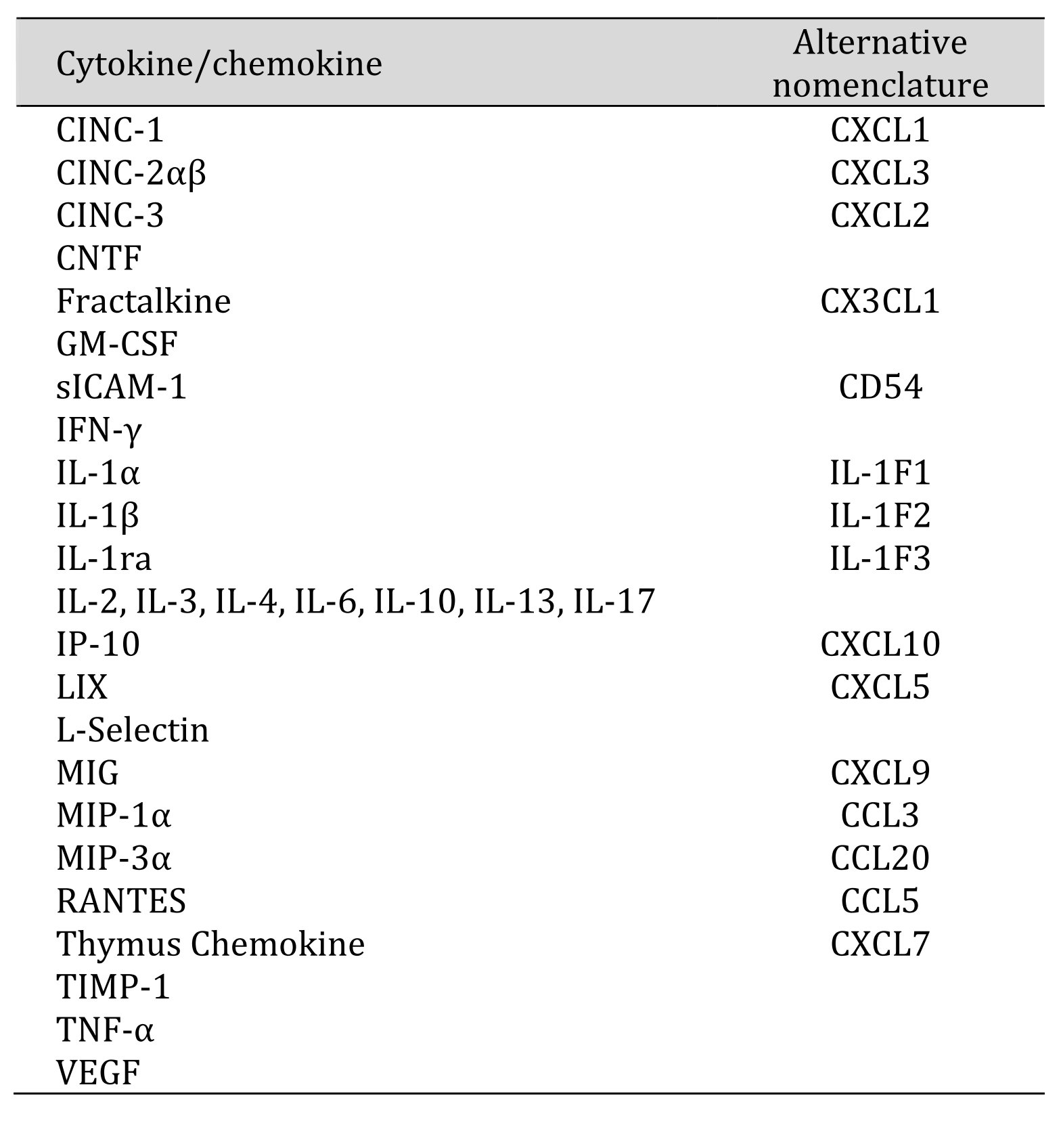
Table 1: The list of proteins detected by the Proteome Profiler Cytokine Array
Western blot assay
Briefly, cells incubated with 20 µM DHA for 12 h and next treated with 10 ng/ml LPS for 2.5 h were lysed with M-PER Reagent supplemented with Protease and Phosphatase Inhibitor Cocktails and the extracts were aliquoted and stored at −80°C. To inhibit Src kinase, before DHA incubation microglia were pretreated for 1h with 1 µM PP2, a reversible, ATP-competitive, inhibitor of the Src family of protein tyrosine kinases. The total protein concentration was measured by Bradford’s method and the concentration was read from a BSA standard curve. Proteins (30 µg total) were separated on SDS-PAGE gels and blotted (120 min, 200 mA, 4°C) onto nitrocellulose membranes (Bio-Rad, USA). The membranes were then blocked for one hour with Tris-buffered saline (TBS) containing 5% BSA at RT and incubated with primary antibodies: anti-TREM21:500 (Novus Biologicals, Inc., USA); anti-Syk 1:1000 (Invitrogen, ThermoFischer Scientific, USA); anti-pSyk 1:1000 (Invitrogen, ThermoFischer Scientific, USA); anti-Akt 1:1000 (Cell Signalling Technology, USA); anti-pAkt 1:2000 (Cell Signalling Technology, USA); anti-GAPDH 1:1200 (Cell Signalling Technology, USA). After overnight at 4 °C, the membranes were washed, incubated for 1.5 h at RT with horseradish peroxidase-conjugated secondary antibodies (Jackson Immunoresearch, USA) (1:10000) and further processed for chemiluminescence detection. The antibody complexes were detected using Pierce ECL Western Blotting Substrate. The images were documented by ChemiDoc MP Imager (BioRad). Densitometric analysis was carried out using ImageLab software ver 6.1. The Western blot analysis was performed in triplicate.
Immunofluorescence phagocytosis assay
Cells plated at density of 120 × 103 cells/well on 1.5-mm2 coverslips were preincubated with 1 µM PP2 for 1h and then exposed to 20 µM DHA for 12 h. Then, fluorescence-labeled latex beads were added (5 μl/ml for 1 h at 37 °C). Cells were washed three times with PBS to remove the non-phagocytized beads, fixed with 4% paraformaldehyde (Avantar Performance Materials, Poland) in PBS at RT and permeabilized with 0.2% Triton X-100 (Avantar Performance Materials, Poland). Subsequently, they were blocked with 2% serum in PBS and incubated overnight at 4 °C with anti-IBA1 antibodies (1:500). Next day, Alexa 594-conjugated secondary antibodies (Thermo Fisher Scientific, USA) were applied at RT, in the dark for 2 h. Finally, the coverslips with the cells were mounted on glass microscope slides using ProLong Gold antifade reagent with DAPI (Thermo Fischer Scientific, USA). Cells with phagocytic activity were analyzed using a AxioExaminer epifluorescence microscope (Carl Zeiss, Germany) equipped with a water immersion objective. Images were captured at 40× magnification. the number of phagocytic cells containing beads (Ncell with beads) was counted in 10 images and presented as a percentage of the total counted cells (Ntotal) for each group. The phagocytic microglia activity was calculated as the number of beads per phagocytic cell. All the experiments were repeated twice.
RNA Isolation and Gene Expression Analysis
The total RNA was isolated using the Trizol reagent (Invitrogen, ThermoFischer Scientific, USA), then purified with the PureLink RNA Mini Kit (Invitrogen, ThermoFischer Scientific, USA) according to the manufacturer’s instructions. The RNA concentration and purity were examined spectrophotometrically in duplicate. From each sample, 1 µg of RNA was reverse-transcribed using M-MLV reverse transcriptase (Promega, USA) and oligo(dT)15 primer (Promega, USA) in a 25 µl reaction volume. The cDNA samples were used for quantitative real-time PCR (qRT-PCR) with specific primers (Table 2 [36–38]). The constitutively expressed GAPDH gene was applied as a reference gene. Quantitative RT-PCR was carried out in Rotorgene 6000 machine (Corbett Research, UK) using KAPA™ SybrFast qPCR Master Mix (Kapa Biosystems, the Netherlands), according to the manufacturer’s instructions, in a total volume of 20 µL and 1/10 cDNA dilution for each tested sample. The annealing temperature for all the primers was 58°C. The melting curves of the amplified products were analyzed at the end of each PCR, and the analysis was carried out at 72-95°C, with temperature increased by 1°C/5s. Four ten-fold dilutions of cDNA were run together with the analyzed samples for a calculation of the standard curve (correlation coefficient > 0.99) and the PCR efficiency. The relative quantification of the mRNA level of the tested genes was read out from the standard curve and normalized to the GAPDH gene. All the calculations were done using the Rotor-Gene 6000 Series Software 1.7 (Corbett Research, United Kingdom). The results were obtained from two independent experiments each in triplicate.
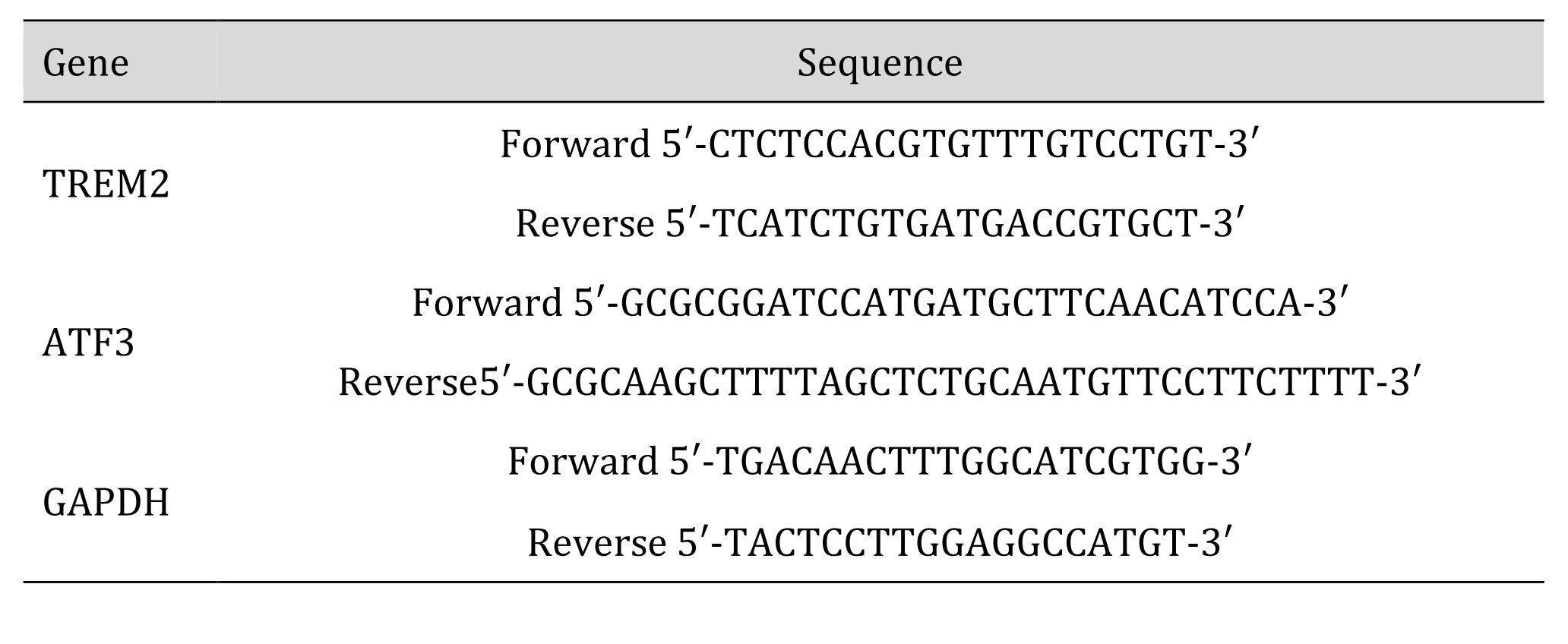
Table 2: Sequences of the primer pairs used for the RT-PCR experiments
Statistical analysis
The results are presented as mean values ± SEM. All the data were analyzed using the GraphPad Prism software 6.0. Statistical significance was determined by one-way ANOVA with Bonferroni correction. For nonparametric data, the Kruskal–Wallis test, followed by the Dunn’s multiple comparison test, was applied. The level of significance was set at p < 0.05.
Results
Viability of microglia treated with DHA and LPS
The results show that 12-hour cell incubation with DHA above 20 µM was increasingly cytotoxic (Fig. 1A). DHA at 30 µM decreased microglia viability to 65.4% and at 50 µM to 35.4%. Therefore, for further experiments DHA at concentration of 20 µM have been chosen. Then we determined dose-dependent decrease in cell viability after LPS treatment (in the range from 2.5 to 1000 ng/ml) (Fig. 1B) and established the working concentration of LPS to 10 ng/ml. Next, we examined whether DHA at a selected concentration was not harmful to LPS-stimulated microglia. Viability of microglia incubated with DHA and treated with 10 ng/ml LPS was similar to LPS-only treated cells and was not significantly different from viability of control cells (Fig. 1C).
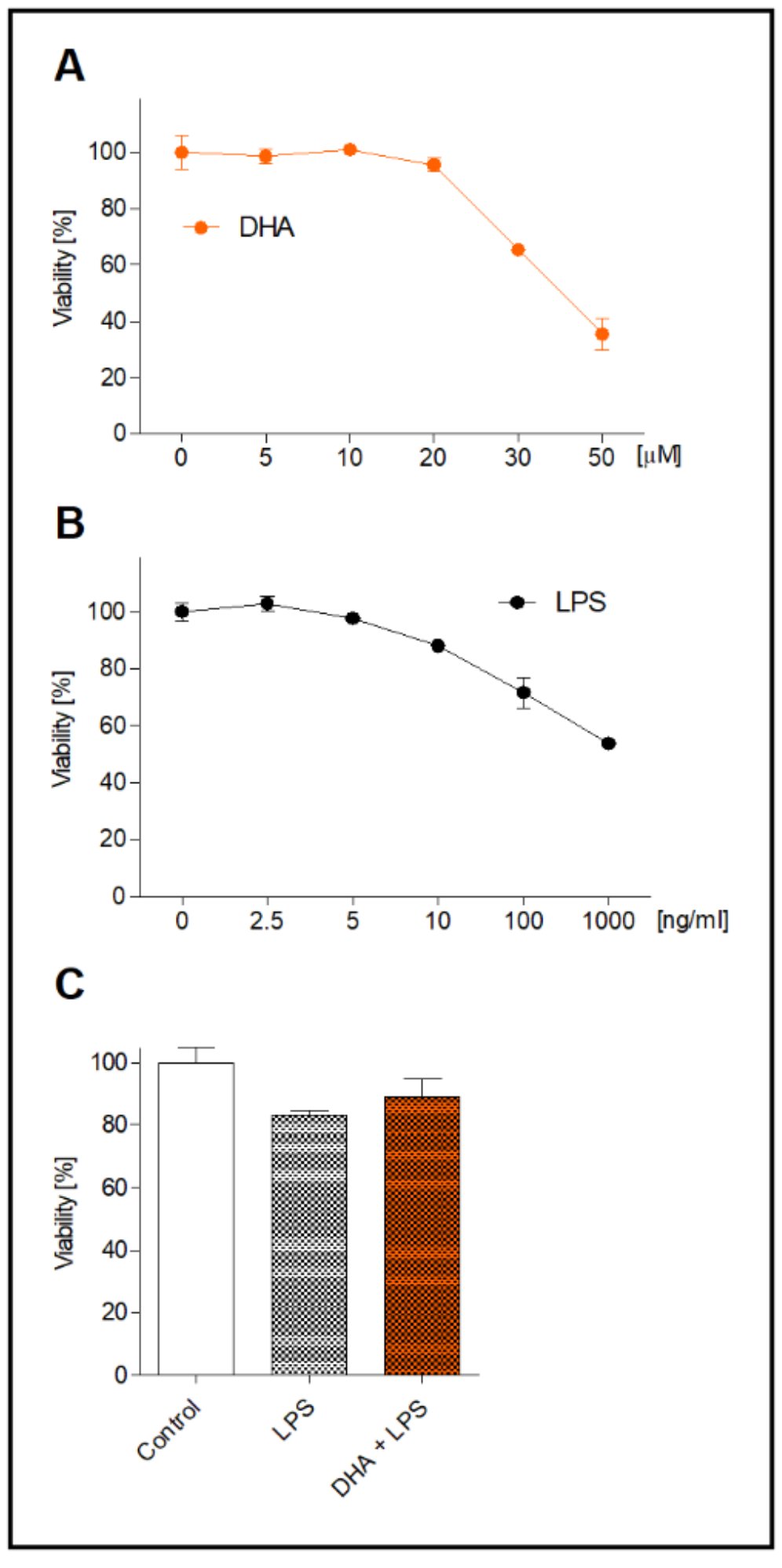
Fig. 1: Viability of microglia incubated with various concentration of DHA for 12 h (A) and LPS for 2.5 h (B). Viability of microglia incubated with 20 µM DHA followed by 10 ng/ml LPS treatment (C). Data is expressed as a mean viability of control cell in percentages ± SEM from three independent experiments.
Effect of DHA on proinflammatory response of LPS-treated microglia
Treatment of microglia with LPS resulted in the expression of proteins not present in non-activated microglia (CINC-1, CINC-2αβ, CINC-3, IL-1α and IL-1β) (Fig. 2A and B). Moreover, the expression of MIP-1α increased 9-fold and the ICAM-1 expression increased by about 30% compared to control cells. Cell preincubation with DHA inhibited the expression of the proinflammatory proteins in activated microglia. The strongest inhibitory effect of DHA in comparison to LPS-treated cells was found for CINC-2αβ (by 68%) and IL-1β (by 48%). The expression of other proteins, i.e., CINC-3, MIP-1α, CINC-1 and IL-1α, was decreased by 30%, 25%, 20% and 16%, respectively (Fig 2B).
In order to determine whether the ATF3 factor, involved in the reduction of inflammatory response, is controlled by DHA, the ATF3 gene expression was assayed. Incubation of resting microglia with DHA increased the ATF3 expression by two-fold (p<0.001) compared to control cells. In LPS-induced microglia, preincubation of cell with DHA significantly increased ATF3 as compared to LPS-treated cells (p<0.05) (Fig. 2C).
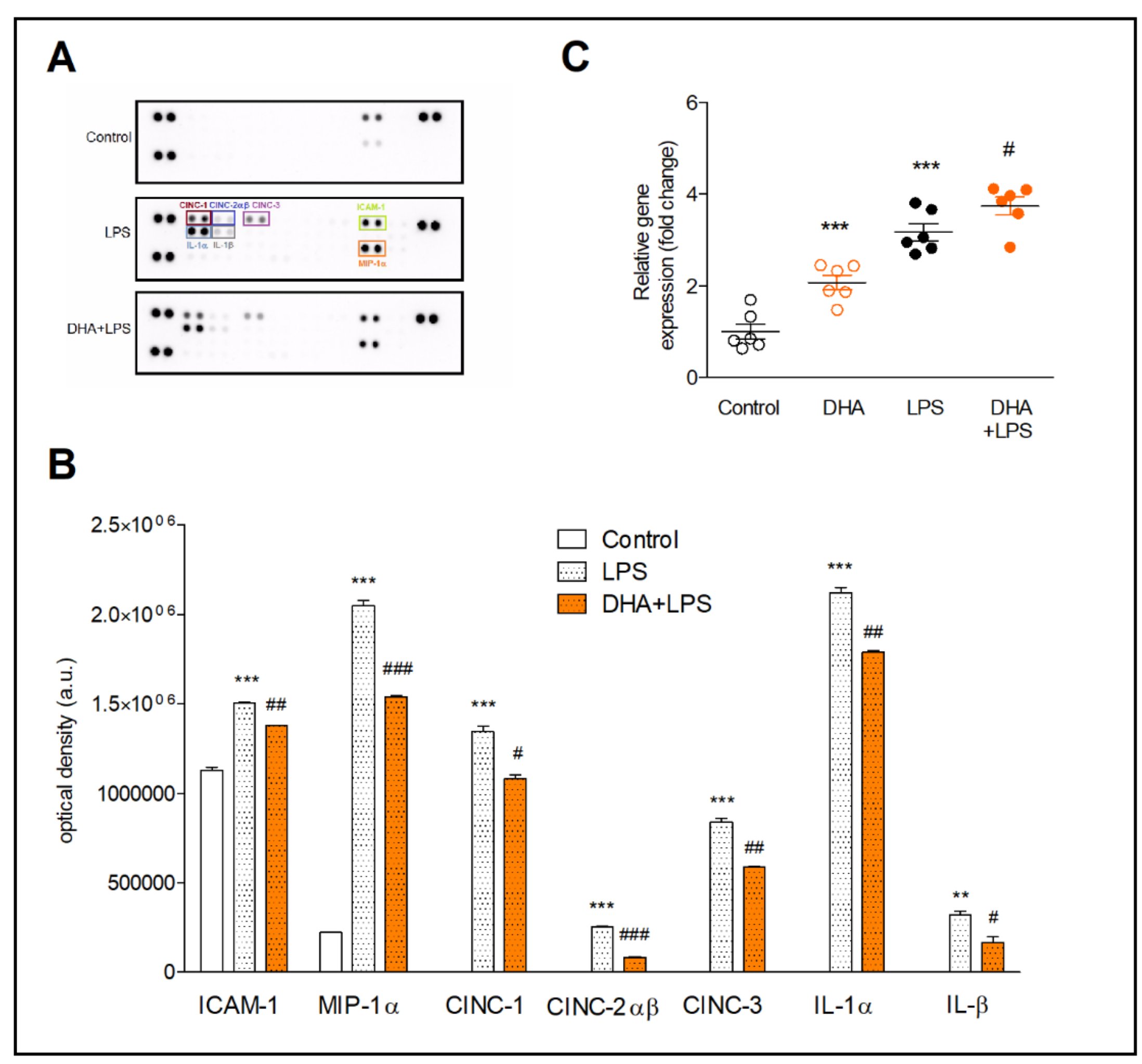
Fig. 2: Proteome profiler arrays (A and B) and expression of ATF3 gene (C) in microglia. Representative array membranes (A) and the relative levels of cytokines and chemokines (B) in microglia preincubated with 20 µM DHA and next treated with 10 ng/ml LPS. Determination of gene and protein expression is described in the method section. The results are given as mean ±SEM. **p<0.01, ***p<0.001 vs control; #p<0.05 ##p<0.01, ###p<0.001 vs LPS-treated cells.
Effect of DHA on TREM2 gene and protein expression
The results of qRT-PCR showed that DHA increased the TREM2 gene expression insignificantly (by approximately 20%) and LPS downregulated TREM2 mRNA by half (p<0.001), both in comparison to control cells (Fig. 3A). The supplementation of microglia with DHA before LPS treatment had no significant effect on TREM2 gene expression.
The Western blot analysis revealed increased TREM2 expression after incubation with DHA activated microglia, however, not significantly. Interestingly, next to 28 kDa molecular weight of TREM2, the additional band of 72 kDa TREM2 appeared on the membrane. This form of TREM2 is most likely the glycosylated TREM2 protein (Fig. 3B).
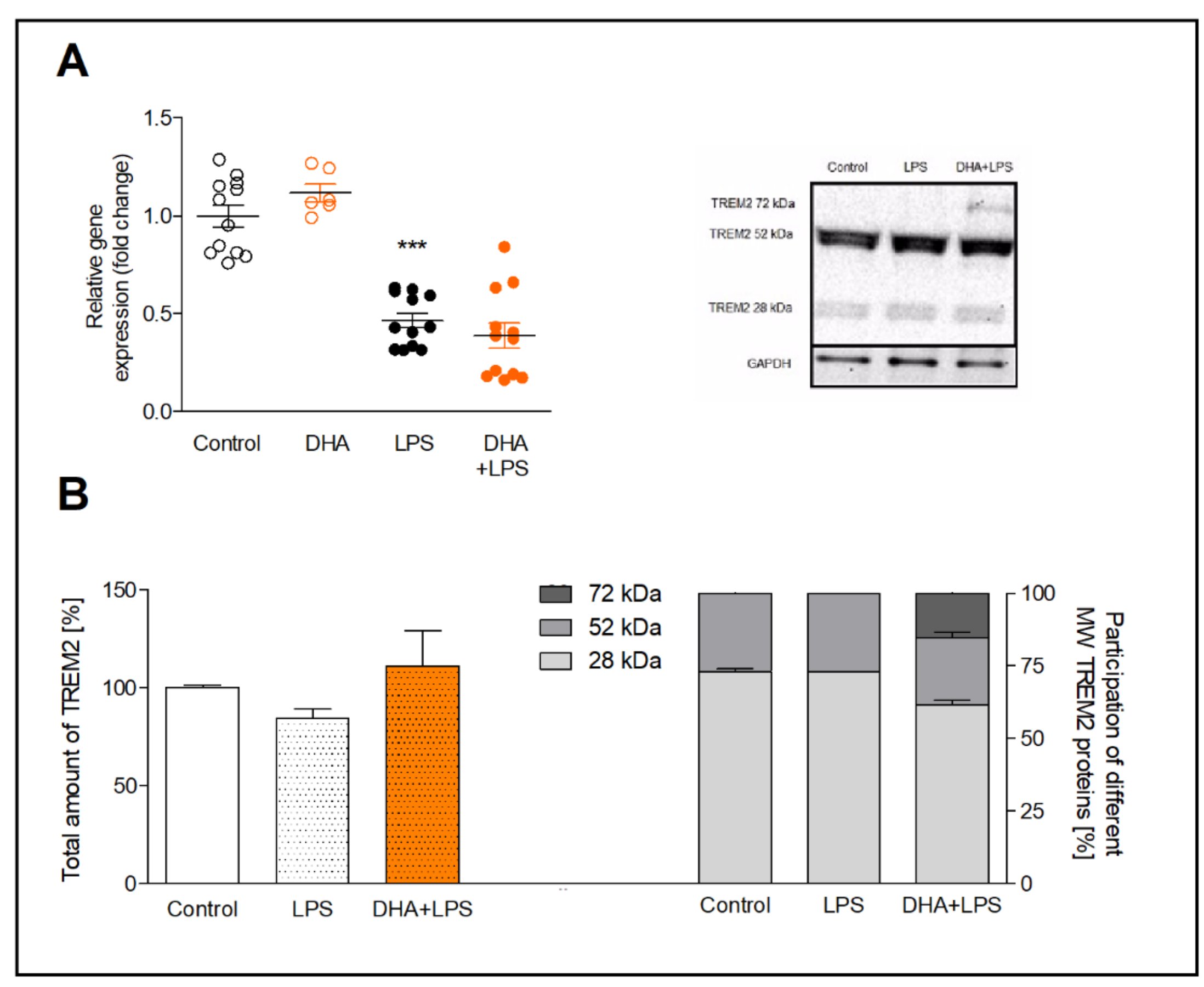
Fig. 3: Expression of TREM2 gene (A) and TREM2 protein (B) in microglia preincubated with DHA and treated with LPS. The WB results are given as a percentage of the total TREM2 level in each group (left bottom graph) and the percentage of each molecular weight form of TREM2 (right bottom graph) with a representative WB blot (right upper image). The results are mean ±SEM from three independent experiments. ***p<0.001 vs control.
Effect of DHA on the activation of kinases downstream of TREM2
We determined the activation of Syk and Akt kinases by measuring their phosphorylated (pSyk, pAkt) and unphosphorylated forms (Syk, Akt) in microglia preincubated with DHA and treated with LPS. The ratio of pAkt/Akt in LPS-treated cells, incubated and not incubated with DHA, did not differ significantly (Fig. 4A). The analysis of Syk kinase activation in LPS-treated microglia showed a slight (11%) decrease in the pSyk/Syk ratio, while DHA pre-treatment of activated cells increased this ratio by 20% (p<0.01) (Fig. 4B)
To examine whether the Syk kinase is activated by DHA through the Src family of tyrosine kinases, we used PP2 reversible inhibitor of Src kinase. In microglia pretreated with PP2 and next incubated with DHA, the pSyk/Syk ratio was 3-fold lower in comparison to cells incubated exclusively with DHA (p<0.001) (Fig. 4C). Such results indicate that DHA induces Syk phosphorylation through activation of the Src kinases.
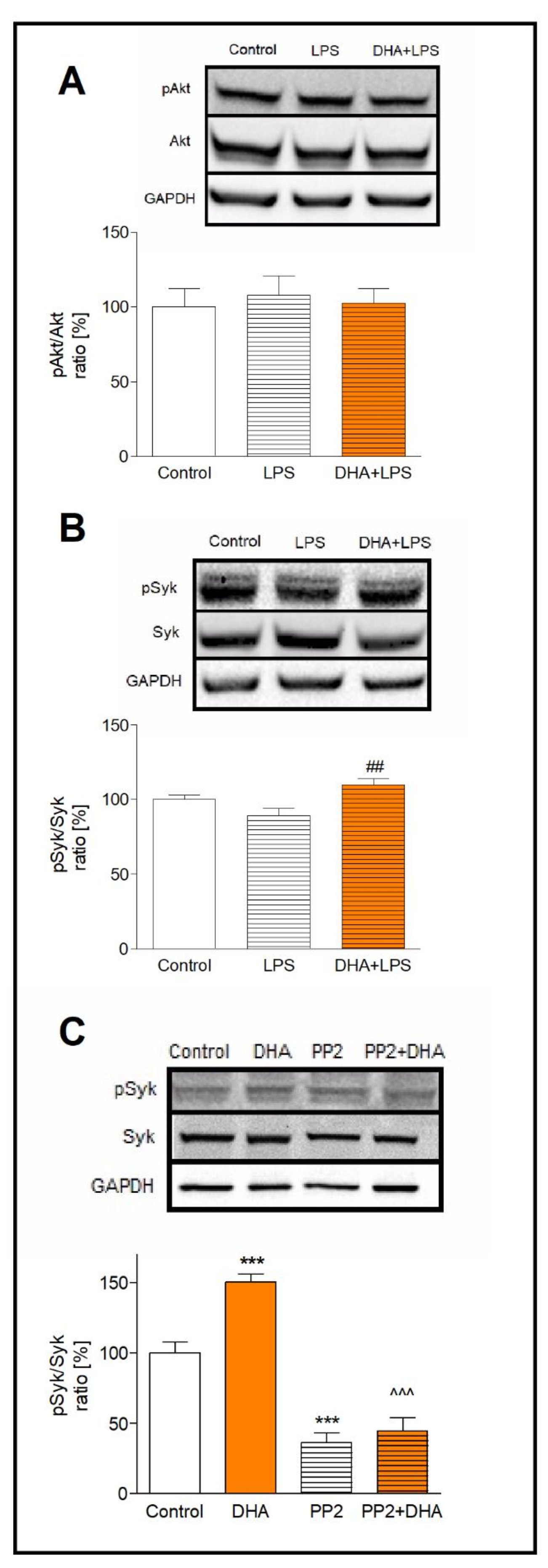
Fig. 4: Activation of Akt and Syk kinases presented as the pAkt/Akt ratio (A) and the pSyk/Syk ratio (B). Activation of Syk kinase in DHA treated microglia after inhibition of the Src kinase by PP2 inhibitor (C). Microglia were treated as described in the method section. The results are given as mean ±SEM from three independent experiments. ***p<0.001 vs control, ##p<0.01 vs LPS-treated cells, ^^^p<0.001 vs DHA-treated cells.
Effect of DHA on microglial phagocytosis
The result of previous works indicate that Syk kinase activation is a key regulator of microglial phagocytosis. Thus, we investigated whether activation of Src/Syk kinases by DHA may increase microglial phagocytosis. We found that a number of the phagocytic cells incubated with DHA, did not differ significantly in comparison to control cells. However, DHA significantly (p<0.05) increased the number of fluorescence-labeled latex beads per phagocytic cell indicating enhanced phagocytic microglia activity. Pretreatment of cells with the Src inhibitor, PP2, completely blocked the beads phagocytosis, and DHA did not reverse this effect, indicating that DHA induces phagocytosis most likely via the Src-dependent pathway (Fig. 5).
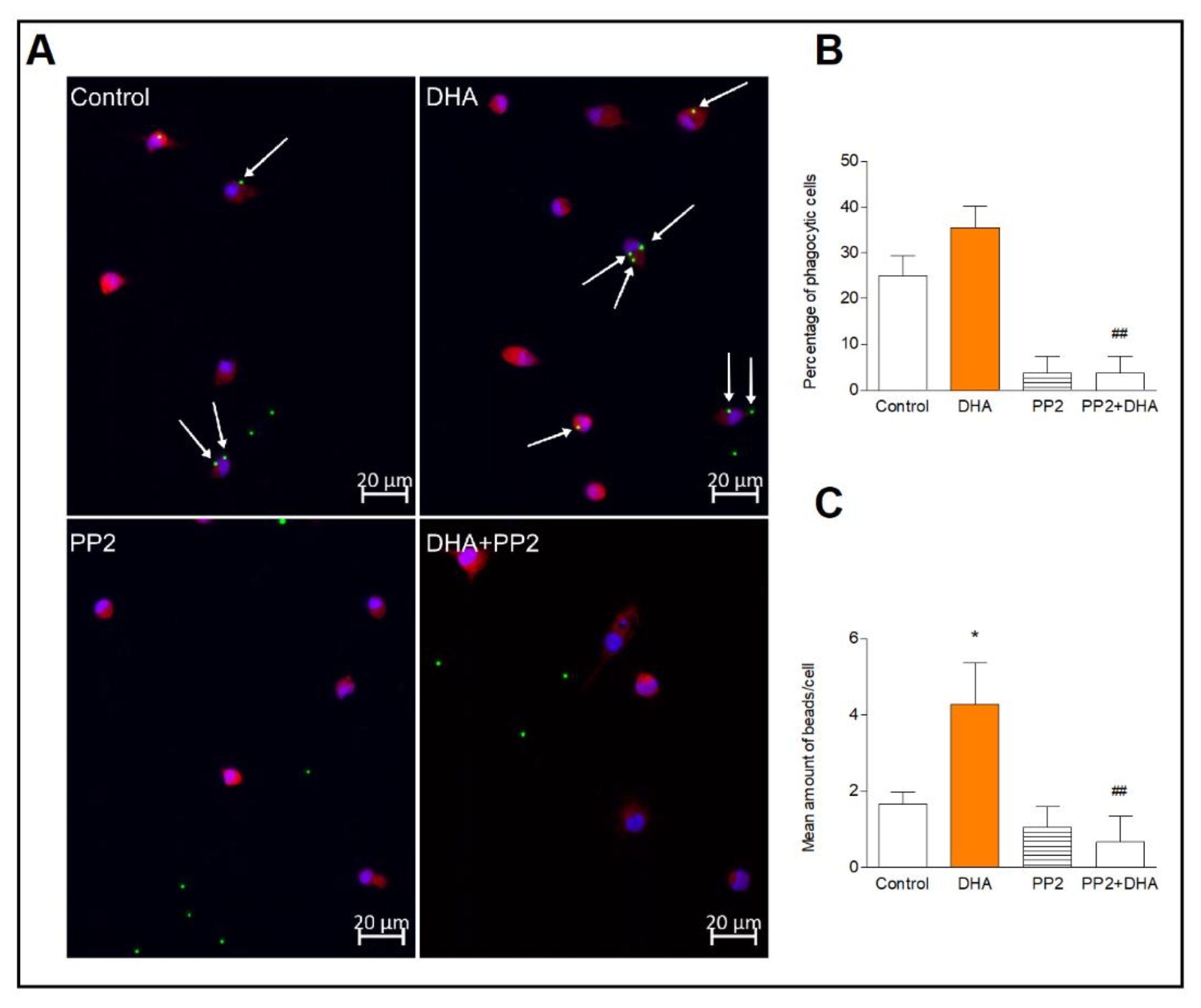
Fig. 5: Phagocytic activity of microglia. Representative images of microglia incubated with fluorescent latex beads (green), immunolabelled for Iba1 to visualize cytoplasmic volume (red) and nuclei with DAPI (blue) (A). Mean percentage of phagocytic cells counted as described in the method section (B). Mean number of beads in phagocytic cells (C). The results are given as mean ±SEM from two independent experiments. *p<0.05 vs control, ##p<0.01 vs DHA-treated cells.
Discussion
In the present study, primary microglia were used to identify the DHA-dependent transcription factor which restrains the inflammatory response of active microglia, and to investigate an activation of the TREM2 receptor downstream kinases, involved in the regulation of microglial phagocytosis. As expected, the treatment of microglia with LPS, markedly increased expression of pro-inflammatory mediators, not expressed in resting microglia, cytokines (IL-1α, IL-1β), chemokines (CINC-1, CINC-2αβ, CINC-3, MIP-1α) and ICAM-1 adhesion receptor. DHA effectively inhibited expression of these pro-inflammatory and chemoattractant protein which corresponds to the results of the study on cortex glial culture [39]. It has also been demonstrated that in rats, after a traumatic brain injury, DHA promoted a shifting of microglia from a pro-inflammatory M1 phenotype to an anti-inflammatory M2 phenotype [40], which helps in resolution of neuroinflammation, a crucial pathogenic hallmark in different neurodegenerative disorders. The present anti-inflammatory DHA effect on the expression of pro-inflammatory cytokines in LPS-induced microglia is consistent with the previous result [41], however, in our study, this effect was observed for a DHA concentration of 20 µM, which was lower than concentration used in the BV-2-immortalized microglia line [41]. The difference in the DHA dose used in our and previous studies, arises most likely from a cell origin, primary microglia vs immortalized BV-2 cells, since BV-2 cells are less fragile to irradiation [42], as well, a lower responsiveness to LPS and IFN-γ stimulation [43]. Furthermore, the immortalized cell lines, such as BV-2 and HAPI, are considered as an inadequate culture system for assessing microglial response to LPS stimulation since they only partially resemble a functional microglia [44].
Previous studies on the anti-inflammatory effect of DHA on microglia showed a reduction of NF-κB activation associated with decreased expression of CD14 and TLR4 cell surface receptors following LPS treatment [41]. Moreover, we have previously demonstrated that DHA increased the activity of NF-κB and AP-1 transcription factors in astrocytes [45, 46]. Another important transcription factor, strongly induced in activated microglia and regulating the expression of cytokines, is ATF3, a member of the CREB/ATF transcription factors family. The gene promoter of ATF3 contains a TATA box and the binding sites for transcription factors such as ATF/CRE, AP-1, p53, E2F and NF-κB [47–49]. We observed that LPS treatment significantly increased the ATF3 expression in microglia. This is in agreement with the previous studies [50–52] and confirms that ATF3 is a negative regulator of the TLR signaling pathway activated in our experiment by LPS [53–55]. A reduction of pro-inflammatory cytokine expression by ATF3 has been reported after middle cerebral artery occlusion in rats [57]. Furthermore, ATF3 KO mice were much more susceptible to endotoxic shock-induced death than their wild-type littermates [58]. Therefore, upregulation of the ATF3 expression by DHA seen in our study support the hypothesis that the anti-inflammatory effect of DHA in resting and LPS-treated microglia, next to a regulation of the transcription factors inhibiting the proinflammatory gene expression, results also from the upregulation of the ATF3 expression.
The TREM2, a transmembrane glycoprotein highly expressed in microglia, inhibits cytokine production in response to TLR activation by ligands [59]. It has also been reported the high expression of TREM2 in microglia surrounding amyloid plaques in APP/PS1 mice [60] and an attenuated pathological inflammation in the brain of mice with TREM2 deficiency [61]. TREM2, activated by various lipid ligands, is described as a phagocytic receptor for micropathogens [62] and promotes phagocytosis of apoptotic neurons [63] and tissue debris [64]. Moreover, previous studies demonstrated that TREM2 mutations resulted in markedly less microglial clustering around Aβ plaques in AD mouse model [25] and are associated with an increased risk of AD development [65]. Though the correlation between the TREM2 and ATF3 expression has been reported [66, 67], in the present study we did not find a change in the TREM2 expression in microglia challenged with DHA. However, an additional 72 kDa protein band was identified that may be related to the glycosylated form of TREM2 [68]. It has been reported that TREM2 mutation impairs N-glycosylation and trafficking of TREM2 from endoplasmic reticulum/Golgi to plasma membrane, thus inhibiting microglial phagocytosis [68, 69]. Nevertheless, a specific mechanism of DHA-induced glycosylation of TREM2 and modification of its subcellular transport to the cell surface remain yet to be elucidated.
The unchanged expression of TREM2, however, was accompanied by increased microglial phagocytosis. TREM2 receptor transmits intracellular signals through two transmembrane adaptors, DAP12 and DAP10, whose phophorylation by the Src family kinases recruits the protein tyrosine kinase Syk, or PI3K, respectively, activating multiple downstream signaling mediators [70, 71]. To explore a possible signaling pathway mediating the DHA effect, our study investigated activation of kinases downstream of TREM2 signaling, Syk and PI3K, a primary Akt kinase activator. We have found, that only the Syk kinase was markedly induced in microglia pretreated with DHA. Additionally, PP2, the Src inhibitor, completely abolished the DHA effect which suggests that DHA acts through activation of Src kinases. The important role of Syk kinase in supporting the CNS function during neuroinflammation has previously been demonstrated in Syk knockout AD model mice and in MS mice [72, 73]. Assuming that the Src/Syk-dependent pathway is involved in phagocytosis regulation [74–76], we conclude that Src kinases can also mediate DHA-dependent enhanced phagocytic microglial activity. A role of the Src kinases in the DHA effect has been demonstrated both in human microglia [34] and N9 microglial cells [77]. Furthermore, since Src-dependent upregulation of the ATF3 expression has been reported [78] a contribution of the Src/Syk dependent upregulation of the ATF3 expression by DHA should be considered.
Conclusion
To summarize, the present study demonstrate that DHA inhibits the expression of cytokines in activated microglia and increases their phagocytic activity, partly through activation of Src/Syk/ATF3 pathway, downstream kinases and transcription factor in TREM2 signalling. Numerous studies based on dietary intervention or survey, reported beneficial effect of n-3 PUFAs in alleviating symptoms of diseases associated with neuroinflammation, such as Alzheimer’s and Parkinson’s diseases [79–82]. We believe that the present findings provide a new insight into understanding of the DHA protective effect in neuroinflammation and may help to develop a new strategy for microglia-activated inflammation.Acknowledgements
The authors are grateful to Barbara Dziedzic for her comments, suggestions and technical assistance and to Tomasz Przygodzki for providing access to the measuring equipment and fluorescence microscope.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Author Contributions
Conceptualization, E.Z.; methodology, E.Z. and. K.W-Sz.; investigation, E.Z., M.M. and. K.W-Sz.; writing—original draft preparation, E.Z., K.W-Sz, A.W., and M.M.; writing—review and editing, E.Z., K.W-Sz, visualization, E.Z.; funding acquisition, E.Z.
Funding Sources
This research was funded by NCN (National Science Centre, Poland) MINIATURA grant no. DEC-2021/05/X/NZ3/01074.
Disclosure Statement
The authors have no conflicts of interest to declare.
References
| 1 | Shigemoto-Mogami Y, Hoshikawa K, Goldman JE, Sekino Y, Sato K. Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J Neurosci Off J Soc Neurosci. 2014 Feb;34(6):2231-43.
https://doi.org/10.1523/JNEUROSCI.1619-13.2014 |
| 2 | Wlodarczyk A, Holtman IR, Krueger M, Yogev N, Bruttger J, Khorooshi R, et al. A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J. 2017 Nov;36(22):3292-308.
https://doi.org/10.15252/embj.201696056 |
| 3 | Hagemeyer N, Hanft K-M, Akriditou M-A, Unger N, Park ES, Stanley ER, et al. Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathol. 2017 Sep;134(3):441-58.
https://doi.org/10.1007/s00401-017-1747-1 |
| 4 | Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR 3rd, Lafaille JJ, et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013 Dec;155(7):1596-609.
https://doi.org/10.1016/j.cell.2013.11.030 |
| 5 | Lim S-H, Park E, You B, Jung Y, Park A-R, Park SG, et al. Neuronal synapse formation induced by microglia and interleukin 10. PLoS One. 2013;8(11):e81218.
https://doi.org/10.1371/journal.pone.0081218 |
| 6 | Lenz KM, Nugent BM, Haliyur R, McCarthy MM. Microglia are essential to masculinization of brain and behavior. J Neurosci Off J Soc Neurosci. 2013 Feb;33(7):2761-72.
https://doi.org/10.1523/JNEUROSCI.1268-12.2013 |
| 7 | Thompson KK, Tsirka SE. The Diverse Roles of Microglia in the Neurodegenerative Aspects of Central Nervous System (CNS) Autoimmunity. Int J Mol Sci. 2017 Feb;18(3). DOI: 10.3390/ijms18030504
https://doi.org/10.3390/ijms18030504 |
| 8 | Kwon HS, Koh S-H. Neuroinflammation in neurodegenerative disorders: the roles of microglia and astrocytes. Transl Neurodegener. 2020 Nov;9(1):42.
https://doi.org/10.1186/s40035-020-00221-2 |
| 9 | Colonna M, Butovsky O. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu Rev Immunol. 2017 Apr;35:441-68.
https://doi.org/10.1146/annurev-immunol-051116-052358 |
| 10 | Guo S, Wang H, Yin Y. Microglia Polarization From M1 to M2 in Neurodegenerative Diseases. Front Aging Neurosci. 2022;14:815347.
https://doi.org/10.3389/fnagi.2022.815347 |
| 11 | Tang Y, Le W. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol Neurobiol. 2016 Mar;53(2):1181-94.
https://doi.org/10.1007/s12035-014-9070-5 |
| 12 | Dhandapani R, Neri M, Bernhard M, Brzak I, Schweizer T, Rudin S, et al. Sustained Trem2 stabilization accelerates microglia heterogeneity and Aβ pathology in a mouse model of Alzheimer's disease. Cell Rep. 2022 May;39(9):110883.
https://doi.org/10.1016/j.celrep.2022.110883 |
| 13 | Schmid CD, Sautkulis LN, Danielson PE, Cooper J, Hasel KW, Hilbush BS, et al. Heterogeneous expression of the triggering receptor expressed on myeloid cells-2 on adult murine microglia. J Neurochem. 2002 Dec;83(6):1309-20.
https://doi.org/10.1046/j.1471-4159.2002.01243.x |
| 14 | Shirotani K, Hori Y, Yoshizaki R, Higuchi E, Colonna M, Saito T, et al. Aminophospholipids are signal-transducing TREM2 ligands on apoptotic cells. Sci Rep. 2019 May;9(1):7508.
https://doi.org/10.1038/s41598-019-43535-6 |
| 15 | Xue T, Ji J, Sun Y, Huang X, Cai Z, Yang J, et al. Sphingosine-1-phosphate, a novel TREM2 ligand, promotes microglial phagocytosis to protect against ischemic brain injury. Acta Pharm Sin B. 2022 Apr;12(4):1885-98.
https://doi.org/10.1016/j.apsb.2021.10.012 |
| 16 | Song W, Hooli B, Mullin K, Jin SC, Cella M, Ulland TK, et al. Alzheimer's disease-associated TREM2 variants exhibit either decreased or increased ligand-dependent activation. Alzheimers Dement. 2017 Apr;13(4):381-7.
https://doi.org/10.1016/j.jalz.2016.07.004 |
| 17 | Stefano L, Racchetti G, Bianco F, Passini N, Gupta RS, Panina Bordignon P, et al. The surface-exposed chaperone, Hsp60, is an agonist of the microglial TREM2 receptor. J Neurochem. 2009 Jul;110(1):284-94.
https://doi.org/10.1111/j.1471-4159.2009.06130.x |
| 18 | Bailey CC, DeVaux LB, Farzan M. The Triggering Receptor Expressed on Myeloid Cells 2 Binds Apolipoprotein E. J Biol Chem. 2015 Oct;290(43):26033-42.
https://doi.org/10.1074/jbc.M115.677286 |
| 19 | Zhao Y, Wu X, Li X, Jiang L-L, Gui X, Liu Y, et al. TREM2 Is a Receptor for β-Amyloid that Mediates Microglial Function. Neuron. 2018 Mar;97(5):1023-1031.e7.
https://doi.org/10.1016/j.neuron.2018.01.031 |
| 20 | Ulland TK, Colonna M. TREM2 - a key player in microglial biology and Alzheimer disease. Nat Rev Neurol. 2018 Nov;14(11):667-75.
https://doi.org/10.1038/s41582-018-0072-1 |
| 21 | Konishi H, Kiyama H. Microglial TREM2/DAP12 Signaling: A Double-Edged Sword in Neural Diseases. Front Cell Neurosci. 2018;12:206.
https://doi.org/10.3389/fncel.2018.00206 |
| 22 | Chouery E, Delague V, Bergougnoux A, Koussa S, Serre J-L, Mégarbané A. Mutations in TREM2 lead to pure early-onset dementia without bone cysts. Hum Mutat. 2008 Sep;29(9):E194-204.
https://doi.org/10.1002/humu.20836 |
| 23 | Ulland TK, Song WM, Huang SC-C, Ulrich JD, Sergushichev A, Beatty WL, et al. TREM2 Maintains Microglial Metabolic Fitness in Alzheimer's Disease. Cell. 2017 Aug;170(4):649-663.e13.
https://doi.org/10.1016/j.cell.2017.07.023 |
| 24 | Jonas LA, Jain T, Li Y-M. Functional insight into LOAD-associated microglial response genes. Open Biol. 2022 Jan;12(1):210280.
https://doi.org/10.1098/rsob.210280 |
| 25 | Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer's disease model. Cell. 2015 Mar;160(6):1061-71.
https://doi.org/10.1016/j.cell.2015.01.049 |
| 26 | Ulrich JD, Finn MB, Wang Y, Shen A, Mahan TE, Jiang H, et al. Altered microglial response to Aβ plaques in APPPS1-21 mice heterozygous for TREM2. Mol Neurodegener. 2014 Jun;9:20.
https://doi.org/10.1186/1750-1326-9-20 |
| 27 | Calder PC. Polyunsaturated fatty acids and inflammation. Biochem Soc Trans. 2005 Apr;33(Pt 2):423-7.
https://doi.org/10.1042/BST0330423 |
| 28 | Delpech J-C, Thomazeau A, Madore C, Bosch-Bouju C, Larrieu T, Lacabanne C, et al. Dietary n-3 PUFAs Deficiency Increases Vulnerability to Inflammation-Induced Spatial Memory Impairment. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol. 2015 Nov;40(12):2774-87.
https://doi.org/10.1038/npp.2015.127 |
| 29 | Yirmiya R, Goshen I. Immune modulation of learning, memory, neural plasticity and neurogenesis. Brain Behav Immun. 2011 Feb;25(2):181-213.
https://doi.org/10.1016/j.bbi.2010.10.015 |
| 30 | Kiecolt-Glaser JK, Epel ES, Belury MA, Andridge R, Lin J, Glaser R, et al. Omega-3 fatty acids, oxidative stress, and leukocyte telomere length: A randomized controlled trial. Brain Behav Immun. 2013 Feb;28:16-24.
https://doi.org/10.1016/j.bbi.2012.09.004 |
| 31 | Wang L, Yuan R, Yao C, Wu Q, Christelle M, Xie W, et al. Effects of resolvin D1 on inflammatory responses and oxidative stress of lipopolysaccharide-induced acute lung injury in mice. Chin Med J (Engl). 2014;127(5):803-9.
|
| 32 | Wang D, Zhang H, Zhang Y, Li W, Sun X, Xing Y, et al [Effects of omega-3 polyunsaturated fatty acids on postoperative inflammatory reaction and clinical efficacy].. Zhonghua Wei Chang Wai Ke Za Zhi. 2015 Jul;18(7):651-5.
|
| 33 | Rey C, Delpech JC, Madore C, Nadjar A, Greenhalgh AD, Amadieu C, et al. Dietary n-3 long chain PUFA supplementation promotes a pro-resolving oxylipin profile in the brain. Brain Behav Immun. 2019 Feb;76:17-27.
https://doi.org/10.1016/j.bbi.2018.07.025 |
| 34 | Hjorth E, Zhu M, Toro VC, Vedin I, Palmblad J, Cederholm T, et al. Omega-3 fatty acids enhance phagocytosis of Alzheimer's disease-related amyloid-β42 by human microglia and decrease inflammatory markers. J Alzheimers Dis. 2013;35(4):697-713.
https://doi.org/10.3233/JAD-130131 |
| 35 | Mizwicki MT, Liu G, Fiala M, Magpantay L, Sayre J, Siani A, et al. 1α,25-dihydroxyvitamin D3 and resolvin D1 retune the balance between amyloid-β phagocytosis and inflammation in Alzheimer's disease patients. J Alzheimers Dis. 2013;34(1):155-70.
https://doi.org/10.3233/JAD-121735 |
| 36 | Jiang X, Zhang J, Xia M, Qiu W, Wang H, Zhao D, et al. Role of activating transcription factor 3 (ATF3) in sublytic C5b-9-induced glomerular mesangial cell apoptosis. Cell Mol Immunol. 2010 Mar;7(2):143-51.
https://doi.org/10.1038/cmi.2009.109 |
| 37 | Zhang L, Dong W, Ma Y, Bai L, Zhang X, Sun C, et al. Pon1 Deficiency Promotes Trem2 Pathway-Mediated Microglial Phagocytosis and Inhibits Pro-inflammatory Cytokines Release In vitro and In vivo. Mol Neurobiol. 2022 Jul;59(7):4612-29.
https://doi.org/10.1007/s12035-022-02827-1 |
| 38 | Claud EC, McDonald JAK, He S-M, Yu Y, Duong L, Sun J, et al. Differential expression of 26S proteasome subunits and functional activity during neonatal development. Biomolecules. 2014 Aug;4(3):812-26.
https://doi.org/10.3390/biom4030812 |
| 39 | Antonietta Ajmone-Cat M, Lavinia Salvatori M, De Simone R, Mancini M, Biagioni S, Bernardo A, et al. Docosahexaenoic acid modulates inflammatory and antineurogenic functions of activated microglial cells. J Neurosci Res. 2012 Mar;90(3):575-87.
https://doi.org/10.1002/jnr.22783 |
| 40 | Chen X, Chen C, Fan S, Wu S, Yang F, Fang Z, et al. Omega-3 polyunsaturated fatty acid attenuates the inflammatory response by modulating microglia polarization through SIRT1-mediated deacetylation of the HMGB1/NF-κB pathway following experimental traumatic brain injury. J Neuroinflammation. 2018 Apr;15(1):116.
https://doi.org/10.1186/s12974-018-1151-3 |
| 41 | De Smedt-Peyrusse V, Sargueil F, Moranis A, Harizi H, Mongrand S, Layé S. Docosahexaenoic acid prevents lipopolysaccharide-induced cytokine production in microglial cells by inhibiting lipopolysaccharide receptor presentation but not its membrane subdomain localization. J Neurochem. 2008 Apr;105(2):296-307.
https://doi.org/10.1111/j.1471-4159.2007.05129.x |
| 42 | Luan W, Li M, Wu C, Shen X, Sun Z. Proteomic dissimilarities of primary microglia and BV2 cells under stimuli. Eur J Neurosci. 2022 Apr;55(7):1709-23.
https://doi.org/10.1111/ejn.15637 |
| 43 | Stohwasser R, Giesebrecht J, Kraft R, Müller EC, Häusler KG, Kettenmann H, et al. Biochemical analysis of proteasomes from mouse microglia: induction of immunoproteasomes by interferon-gamma and lipopolysaccharide. Glia. 2000 Feb;29(4):355-65.
https://doi.org/10.1002/(SICI)1098-1136(20000215)29:4<355::AID-GLIA6>3.0.CO;2-4 |
| 44 | Horvath RJ, Nutile-McMenemy N, Alkaitis MS, Deleo JA. Differential migration, LPS-induced cytokine, chemokine, and NO expression in immortalized BV-2 and HAPI cell lines and primary microglial cultures. J Neurochem. 2008 Oct;107(2):557-69.
https://doi.org/10.1111/j.1471-4159.2008.05633.x |
| 45 | Zgórzyńska E, Dziedzic B, Gorzkiewicz A, Stulczewski D, Bielawska K, Su K-P, et al. Omega-3 polyunsaturated fatty acids improve the antioxidative defense in rat astrocytes via an Nrf2-dependent mechanism. Pharmacol Rep. 2017 Oct;69(5):935-42.
https://doi.org/10.1016/j.pharep.2017.04.009 |
| 46 | Zgorzynska E, Stulczewski D, Dziedzic B, Su K-P, Walczewska A. Docosahexaenoic fatty acid reduces the pro-inflammatory response induced by IL-1β in astrocytes through inhibition of NF-κB and AP-1 transcription factor activation. BMC Neurosci. 2021 Jan;22(1):4.
https://doi.org/10.1186/s12868-021-00611-w |
| 47 | Tamura K, Hua B, Adachi S, Guney I, Kawauchi J, Morioka M, et al. Stress response gene ATF3 is a target of c-myc in serum-induced cell proliferation. EMBO J. 2005 Jul;24(14):2590-601.
https://doi.org/10.1038/sj.emboj.7600742 |
| 48 | Huo JS, McEachin RC, Cui TX, Duggal NK, Hai T, States DJ, et al. Profiles of growth hormone (GH)-regulated genes reveal time-dependent responses and identify a mechanism for regulation of activating transcription factor 3 by GH. J Biol Chem. 2006 Feb;281(7):4132-41.
https://doi.org/10.1074/jbc.M508492200 |
| 49 | Hunt D, Raivich G, Anderson PN. Activating transcription factor 3 and the nervous system. Front Mol Neurosci. 2012;5:7.
https://doi.org/10.3389/fnmol.2012.00007 |
| 50 | Lai P-F, Cheng C-F, Lin H, Tseng T-L, Chen H-H, Chen S-H. ATF3 Protects against LPS-Induced Inflammation in Mice via Inhibiting HMGB1 Expression. Evid Based Complement Alternat Med. 2013;2013:716481.
https://doi.org/10.1155/2013/716481 |
| 51 | Das A, Kim SH, Arifuzzaman S, Yoon T, Chai JC, Lee YS, et al. Transcriptome sequencing reveals that LPS-triggered transcriptional responses in established microglia BV2 cell lines are poorly representative of primary microglia. J Neuroinflammation. 2016 Jul;13(1):182.
https://doi.org/10.1186/s12974-016-0644-1 |
| 52 | Akbarpour Arsanjani A, Abuei H, Behzad-Behbahani A, Bagheri Z, Arabsolghar R, Farhadi A. Activating transcription factor 3 inhibits NF‑κB p65 signaling pathway and mediates apoptosis and cell cycle arrest in cervical cancer cells. Infect Agent Cancer. 2022 Dec;17(1):62.
https://doi.org/10.1186/s13027-022-00475-7 |
| 53 | Whitmore MM, Iparraguirre A, Kubelka L, Weninger W, Hai T, Williams BRG. Negative regulation of TLR-signaling pathways by activating transcription factor-3. J Immunol. 2007 Sep;179(6):3622-30.
https://doi.org/10.4049/jimmunol.179.6.3622 |
| 54 | Rosenberger CM, Clark AE, Treuting PM, Johnson CD, Aderem A. ATF3 regulates MCMV infection in mice by modulating IFN-gamma expression in natural killer cells. Proc Natl Acad Sci U S A. 2008 Feb;105(7):2544-9.
https://doi.org/10.1073/pnas.0712182105 |
| 55 | Labzin LI, Schmidt S V, Masters SL, Beyer M, Krebs W, Klee K, et al. ATF3 Is a Key Regulator of Macrophage IFN Responses. J Immunol. 2015 Nov;195(9):4446-55.
https://doi.org/10.4049/jimmunol.1500204 |
| 56 | Kim K-H, Jeong J-Y, Surh Y-J, Kim K-W. Expression of stress-response ATF3 is mediated by Nrf2 in astrocytes. Nucleic Acids Res. 2010 Jan;38(1):48-59.
https://doi.org/10.1093/nar/gkp865 |
| 57 | Ma N, Li G, Fu X. Protective role of activating transcription factor 3 against neuronal damage in rats with cerebral ischemia. Brain Behav. 2022 Apr;12(4):e2522.
https://doi.org/10.1002/brb3.2522 |
| 58 | Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006 May;441(7090):173-8.
https://doi.org/10.1038/nature04768 |
| 59 | Hickman SE, El Khoury J. TREM2 and the neuroimmunology of Alzheimer's disease. Biochem Pharmacol. 2014 Apr;88(4):495-8.
https://doi.org/10.1016/j.bcp.2013.11.021 |
| 60 | Neumann H, Daly MJ. Variant TREM2 as risk factor for Alzheimer's disease. N Engl J Med. 2013 Jan;368(2):182-4.
https://doi.org/10.1056/NEJMe1213157 |
| 61 | Jay TR, Miller CM, Cheng PJ, Graham LC, Bemiller S, Broihier ML, et al. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer's disease mouse models. J Exp Med. 2015 Mar;212(3):287-95.
https://doi.org/10.1084/jem.20142322 |
| 62 | Neumann H, Takahashi K. Essential role of the microglial triggering receptor expressed on myeloid cells-2 (TREM2) for central nervous tissue immune homeostasis. J Neuroimmunol. 2007 Mar;184(1-2):92-9.
https://doi.org/10.1016/j.jneuroim.2006.11.032 |
| 63 | Takahashi K, Rochford CDP, Neumann H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J Exp Med. 2005 Feb;201(4):647-57.
https://doi.org/10.1084/jem.20041611 |
| 64 | Takahashi K, Prinz M, Stagi M, Chechneva O, Neumann H. TREM2-transduced myeloid precursors mediate nervous tissue debris clearance and facilitate recovery in an animal model of multiple sclerosis. PLoS Med. 2007 Apr;4(4):e124.
https://doi.org/10.1371/journal.pmed.0040124 |
| 65 | Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson P V, Snaedal J, et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med. 2013 Jan;368(2):107-16.
https://doi.org/10.1056/NEJMoa1211103 |
| 66 | Weigelt K, Carvalho LA, Drexhage RC, Wijkhuijs A, de Wit H, van Beveren NJM, et al. TREM-1 and DAP12 expression in monocytes of patients with severe psychiatric disorders. EGR3, ATF3 and PU.1 as important transcription factors. Brain Behav Immun. 2011 Aug;25(6):1162-9.
https://doi.org/10.1016/j.bbi.2011.03.006 |
| 67 | Hou J, Chen Y, Grajales-Reyes G, Colonna M. TREM2 dependent and independent functions of microglia in Alzheimer's disease. Mol Neurodegener. 2022;17(1):84.
https://doi.org/10.1186/s13024-022-00588-y |
| 68 | Park J-S, Ji IJ, An HJ, Kang M-J, Kang S-W, Kim D-H, et al. Disease-Associated Mutations of TREM2 Alter the Processing of N-Linked Oligosaccharides in the Golgi Apparatus. Traffic. 2015 May;16(5):510-8.
https://doi.org/10.1111/tra.12264 |
| 69 | Kleinberger G, Yamanishi Y, Suárez-Calvet M, Czirr E, Lohmann E, Cuyvers E, et al. TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci Transl Med. 2014 Jul;6(243):243ra86.
https://doi.org/10.1126/scitranslmed.3009093 |
| 70 | Xing J, Titus AR, Humphrey MB. The TREM2-DAP12 signaling pathway in Nasu-Hakola disease: a molecular genetics perspective. Res reports Biochem. 2015;5:89-100.
https://doi.org/10.2147/RRBC.S58057 |
| 71 | Peng Q, Malhotra S, Torchia JA, Kerr WG, Coggeshall KM, Humphrey MB. TREM2- and DAP12-dependent activation of PI3K requires DAP10 and is inhibited by SHIP1. Sci Signal. 2010 May;3(122):ra38.
https://doi.org/10.1126/scisignal.2000500 |
| 72 | Wang S, Sudan R, Peng V, Zhou Y, Du S, Yuede CM, et al. TREM2 drives microglia response to amyloid-β via SYK-dependent and -independent pathways. Cell. 2022 Oct;185(22):4153-4169.e19.
https://doi.org/10.1016/j.cell.2022.09.033 |
| 73 | Ennerfelt H, Frost EL, Shapiro DA, Holliday C, Zengeler KE, Voithofer G, et al. SYK coordinates neuroprotective microglial responses in neurodegenerative disease. Cell. 2022 Oct;185(22):4135-4152.e22.
https://doi.org/10.1016/j.cell.2022.09.030 |
| 74 | Korade-Mirnics Z, Corey SJ. Src kinase-mediated signaling in leukocytes. J Leukoc Biol. 2000 Nov;68(5):603-13.
https://doi.org/10.1189/jlb.68.5.603 |
| 75 | Majeed M, Caveggion E, Lowell CA, Berton G. Role of Src kinases and Syk in Fcgamma receptor-mediated phagocytosis and phagosome-lysosome fusion. J Leukoc Biol. 2001 Nov;70(5):801-11.
https://doi.org/10.1189/jlb.70.5.801 |
| 76 | Berton G, Mócsai A, Lowell CA. Src and Syk kinases: key regulators of phagocytic cell activation. Trends Immunol. 2005 Apr;26(4):208-14.
https://doi.org/10.1016/j.it.2005.02.002 |
| 77 | Tremblay M-E, Zhang I, Bisht K, Savage JC, Lecours C, Parent M, et al. Remodeling of lipid bodies by docosahexaenoic acid in activated microglial cells. J Neuroinflammation. 2016;13(1):116.
https://doi.org/10.1186/s12974-016-0580-0 |
| 78 | Nguyen TT, Johnsen IB, Knetter CF, Drabløs F, Fitzgerald KA, Lien E, et al. Differential gene expression downstream of Toll-like receptors (TLRs): role of c-Src and activating transcription factor 3 (ATF3). J Biol Chem. 2010 May;285(22):17011-9.
https://doi.org/10.1074/jbc.M109.068817 |
| 79 | Heneka MT, Carson MJ, El Khoury J, Landreth GE, Brosseron F, Feinstein DL, et al. Neuroinflammation in Alzheimer's disease. Lancet Neurol. 2015 Apr;14(4):388-405.
https://doi.org/10.1016/S1474-4422(15)70016-5 |
| 80 | More SV, Kumar H, Kim IS, Song S-Y, Choi D-K. Cellular and molecular mediators of neuroinflammation in the pathogenesis of Parkinson's disease. Mediators Inflamm. 2013;2013:952375.
https://doi.org/10.1155/2013/952375 |
| 81 | De Virgilio A, Greco A, Fabbrini G, Inghilleri M, Rizzo MI, Gallo A, et al. Parkinson's disease: Autoimmunity and neuroinflammation. Autoimmun Rev. 2016 Oct;15(10):1005-11.
https://doi.org/10.1016/j.autrev.2016.07.022 |
| 82 | Meraz-Ríos MA, Toral-Rios D, Franco-Bocanegra D, Villeda-Hernández J, Campos-Peña V. Inflammatory process in Alzheimer's Disease. Front Integr Neurosci. 2013;7:59.
https://doi.org/10.3389/fnint.2013.00059 |