Association of IL4 (Rs2243250) Gene Variant and Mycoplasma Pneumoniae Infection with Asthma Susceptibility in an Iraqi Population
bTechnical Institute of Babylon, Al-Furat Al-Awsat Technical University (ATU), Iraq,
cBiology Department, College of Science, Al-Qasim Green University, Babylon, Iraq,
dDepartment of Clinical Chemistry, Faculty of Medicine Hammurabi, University of Babylon, Babylon, Iraq
Keywords
Abstract
Background/Aims:
Asthma is a multifactorial disease influenced by both genetic and environmental factors. This study aimed to investigate the association between the IL4 gene polymorphism (rs2243250) and asthma susceptibility, along with serum IL-4 levels. Additionally, it explored Mycoplasma pneumoniae infection as a potential risk factor for asthma.Methods:
A total of 118 individuals were enrolled, including 60 asthma patients and 58 healthy controls. Genotyping for IL4 rs2243250 was performed using allele-specific PCR (AS-PCR). Previous Mycoplasma pneumoniae infection was assessed serologically, and serum IL-4 levels were measured using ELISA.Results:
No significant differences were observed between groups in terms of age, sex, or residence. Smoking (OR: 7.85, P = 0.001) and family history of asthma (OR: 5.33, P = 0.004) were identified as significant risk factors. Mycoplasma pneumoniae infection was significantly more prevalent in asthma patients (41.7%) than in controls, with a strong association with asthma risk (OR: 8.75, P < 0.0001). Genotype frequencies of rs2243250 differed significantly: CC (36.7% vs. 68.9%), CT (41.7% vs. 24.2%), and TT (21.6% vs. 6.9%) in patients versus controls, respectively. The T allele was more frequent among patients (42.5%) than controls (18.97%), increasing asthma risk (OR: 3.16, P = 0.0001). Both CT (OR: 3.25) and TT (OR: 5.91) genotypes were strongly associated with asthma. Moreover, individuals with the TT genotype had significantly higher serum IL-4 levels (P < 0.001).Conclusion:
The IL4 rs2243250 polymorphism is associated with increased asthma susceptibility and elevated serum IL-4 levels in the Iraqi population. Mycoplasma pneumoniae infection also appears to be a significant contributing factor. Larger-scale studies are warranted to confirm these findings and further explore the role of this infection in asthma pathogenesis.Introduction
Asthma is a prevalent, complex, and chronic inflammatory disease that affects individuals of all ages and imposes a considerable burden on healthcare systems [1]. It is characterized by variable airflow limitation and persistent airway inflammation, manifesting as intermittent respiratory symptoms such as wheezing, shortness of breath, chest tightness, and coughing [2]. Globally, asthma affects approximately 300 million people, resulting in an estimated 250, 000 deaths annually [3] .By 2025, this number is projected to rise to 400 million cases [4]. In Iraq, asthma accounts for approximately 200, 000 hospitalizations or emergency department visits each year. Neighboring countries report prevalence rates of 5.6% in Saudi Arabia and 8.5% in Kuwait [5, 6].
Asthma arises from a combination of genetic susceptibility to atopy and increased bronchial responsiveness to environmental stimuli. This pathophysiological process involves the activation of airway cells—including eosinophils, T cells, mast cells, macrophages, neutrophils, epithelial cells, fibroblasts, and bronchial smooth muscle cells—which release various pro-inflammatory cytokines and mediators [7, 8]. Numerous risk factors contribute to asthma development, including genetic predisposition, maternal smoking during pregnancy, prenatal nutrition, antibiotic use, mode of delivery, family size and birth order, exposure to environmental smoke, socioeconomic status, viral infections [9], and certain occupational exposures [10].
The etiology of asthma is multifactorial, involving genetic, environmental, and immunological components. Infections caused by Mycoplasma pneumoniae (Mp), a pathogen commonly associated with pneumonia and bronchitis, have been implicated in the initiation and exacerbation of asthma, particularly in genetically predisposed individuals [11, 12].
Interleukins 4 (IL-4) and 13 (IL-13) are key mediators in type 2 asthma, although their roles are complex. IL-4 promotes T-helper 2 (Th2) cell differentiation, immunoglobulin class switching, and eosinophil recruitment. IL-13 acts synergistically with IL-4 to enhance IgE production, induce nitric oxide synthesis, and drive airway remodeling involving goblet cells, fibroblasts, and smooth muscle cells. Both cytokines activate shared signaling pathways by binding to receptor complexes containing the IL-4Rα subunit, triggering receptor dimerization and downstream pathological effects [13].
Asthma’s clinical presentation, severity, and treatment response vary widely, partly due to underlying genetic polymorphisms. Single nucleotide polymorphisms (SNPs), in particular, contribute to this variability. Understanding these genetic predispositions is crucial for improving asthma management. Advances in asthma genetics have enhanced our knowledge of the disease’s pathogenesis, diagnosis, and potential therapeutic targets [14].
Among the genes implicated in asthma susceptibility, IL-4 has been extensively studied. However, findings regarding the association between IL-4 gene polymorphisms and asthma risk remain inconsistent [15, 16]. The IL-4 gene is located on chromosome 5q31–q33 [17], and its promoter variant C-590T (rs2243250) has been suggested to increase gene expression, elevate plasma IgE levels, and worsen asthma symptoms.
This study aims to investigate the association between the IL-4 rs2243250 polymorphism and asthma susceptibility and to evaluate its impact on serum IL-4 levels in individuals with allergic asthma. Additionally, we explore the potential role of prior Mycoplasma pneumoniae infection as a contributing risk factor for asthma.
Materials and Methods
Study Design and Participants
This case-control study was conducted from January to August 2021 at the Chest and Respiratory Disease Center in Al-Hillah, Babylon, Iraq. A total of 60 patients with clinically diagnosed asthma (20 males and 40 females), aged 20–64 years, were recruited. The diagnosis was confirmed by consultant pulmonologists according to internationally accepted criteria. The control group consisted of 58 age- and sex-matched healthy individuals (25 males and 33 females) with no history of asthma, chronic respiratory disease, autoimmune conditions, or recent respiratory infections. Controls were recruited from the general population and hospital staff after completing a health screening questionnaire. Participants who had received immunosuppressive therapy or antibiotics within the previous month were excluded from both groups. The sample size was determined based on the availability of eligible participants during the study period, as well as by referencing similar previous studies in the field.
Blood and Serum Collection
Five milliliters of venous blood were collected from each participant under aseptic conditions. Of this, three milliliters were placed in plain tubes for serum separation, while two milliliters were collected in EDTA tubes for DNA extraction. All samples were stored at –20 °C until analysis to preserve integrity and ensure reliable laboratory results.
Measurement of IL-4 Cytokine
Serum levels of interleukin-4 (IL-4) were measured using an enzyme-linked immunosorbent assay (ELISA) with a commercial kit (Diaclone, France), following the manufacturers protocol.
Detection of Previous Mycoplasma pneumoniae Infection
To evaluate prior exposure to Mycoplasma pneumoniae, serum IgG levels were assessed using the Mycoplasma pneumoniae IgG Human ELISA Kit (Abcam, UK). According to the manufacturer’s guidelines, samples with IgG concentrations above 10 U/mL were considered seropositive, indicating past infection.
SNP Genotyping
Genomic DNA was extracted from whole blood using the Qiagen DNA extraction kit, following the manufacturer’s protocol. DNA concentration and purity were assessed by measuring absorbance at 260/280 nm using a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA). DNA samples were stored at –20 °C until genotyping analysis.
Allele-specific PCR (AS-PCR) was used to detect the IL-4 gene polymorphism rs2243250, with primers designed according to [18] as detailed in Table 1 .The AS-PCR protocol followed the method described by [19]. Each 25 µL PCR reaction contained 12.5 µL of Master Mix (Promega, USA), 1 µL of each primer, 30 ng of genomic DNA, and nuclease-free water. PCR amplification was performed using a Bio-Rad thermal cycler under the following conditions: initial denaturation at 95 °C for 2 minutes, followed by 30 cycles of denaturation at 95 °C for 30 seconds, annealing at 65 °C for 50 seconds, and extension at 72 °C for 30 seconds, with a final extension at 72 °C for 5 minutes. Amplified products were visualized on a 1.5% agarose gel (Promega, USA) stained with ethidium bromide.
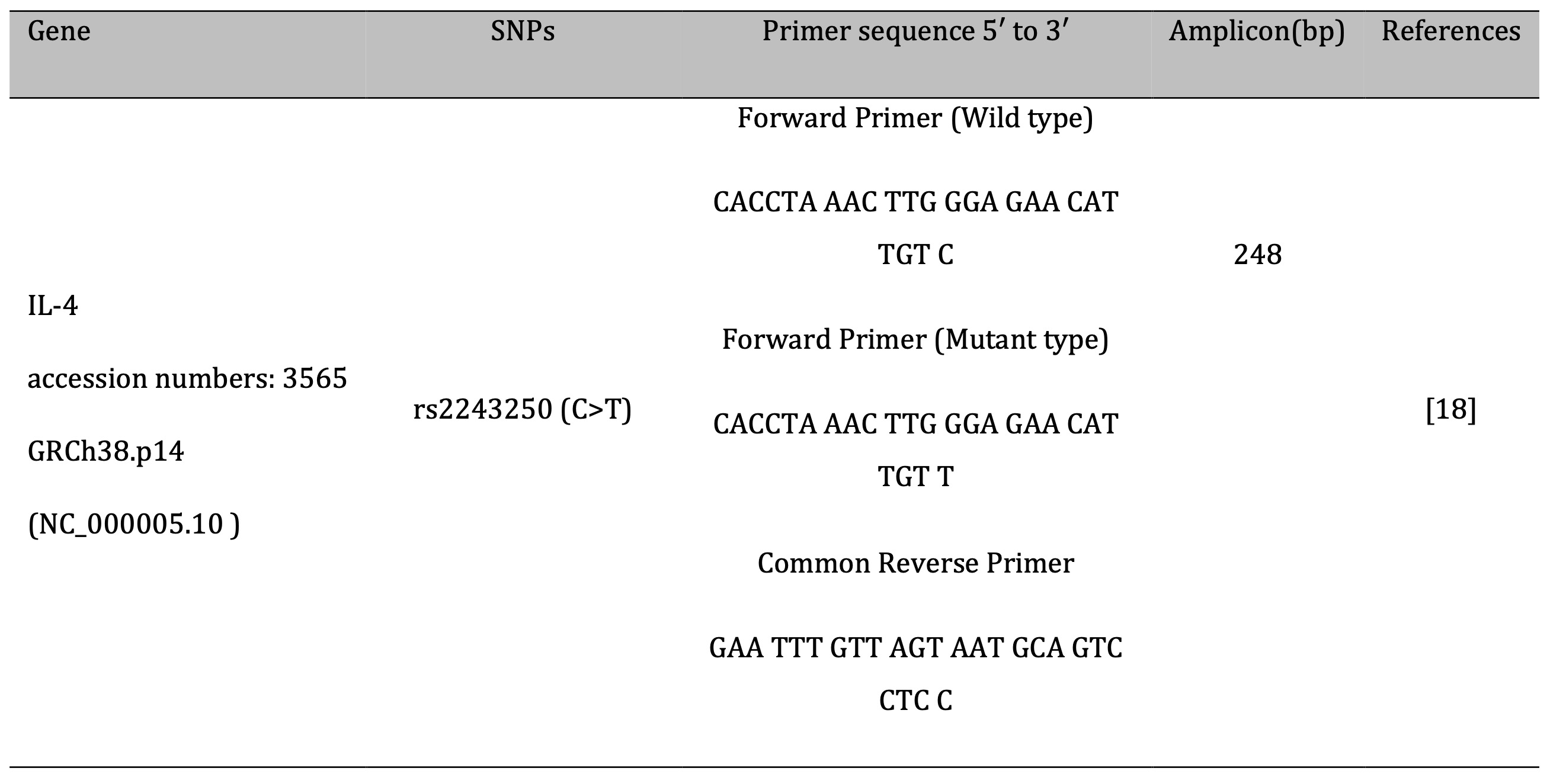
Table 1: Primers used in the study
Statistical Analysis
Serum cytokine levels are presented as mean ± standard deviation (SD). Data normality was assessed before conducting one-way ANOVA to compare group means. Genotype and allele frequencies were calculated by direct counting, and deviations from Hardy–Weinberg equilibrium (HWE) were evaluated using Pearson’s chi-squared test. Genotype distributions are expressed as percentages. Associations between SNP genotypes and asthma risk were analyzed using odds ratios (ORs) with 95% confidence intervals (CIs). A p-value < 0.05 was considered statistically significant. Statistical analyses were performed using SPSS version 23, WinPepi version 11.65, and an online HWE calculator.
Results
Demographic Characteristics and Risk Factors Associated with Bronchial Asthma
The study included 60 patients diagnosed with bronchial asthma and 58 healthy controls. Demographic data are presented in Table 2. The mean age did not differ significantly between the patient group (40.36 ± 11.10 years) and the control group (42.02 ± 10.53 years; P = 0.642). Among patients, 20 (33.3%) were male and 40 (66.7%) were female, while the control group comprised 25 males (43.1%) and 33 females (56.9%). No statistically significant differences were found between the groups in terms of age (P = 0.642), gender (P = 0.274), or place of residence (P = 0.474), indicating proper matching for a case-control design. However, smoking and a positive family history of asthma were identified as significant risk factors. Smokers were 7.85 times more likely to develop asthma (OR: 2.17–28.44; P = 0.001), and individuals with a family history of asthma were 5.33 times more likely to be affected (OR: 1.67–17.03; P = 0.004) as shown in Table 3.
The association between prior Mycoplasma pneumoniae infection and disease status was assessed in both patients and controls, as shown in Fig. 1. The figure demonstrates that M. pneumoniae infection was significantly more prevalent among patients (25/60, 41.7%) compared to controls (8/58, 13.8%). The calculated odds ratio was 8.75 (95% CI: 3.53–21.64; p < 0.0001; Table 4), indicating a strong and statistically significant association between M. pneumoniae infection and the patient group.

Fig. 1: Distribution of Previous Infection of M.pneumoniae among study groups.
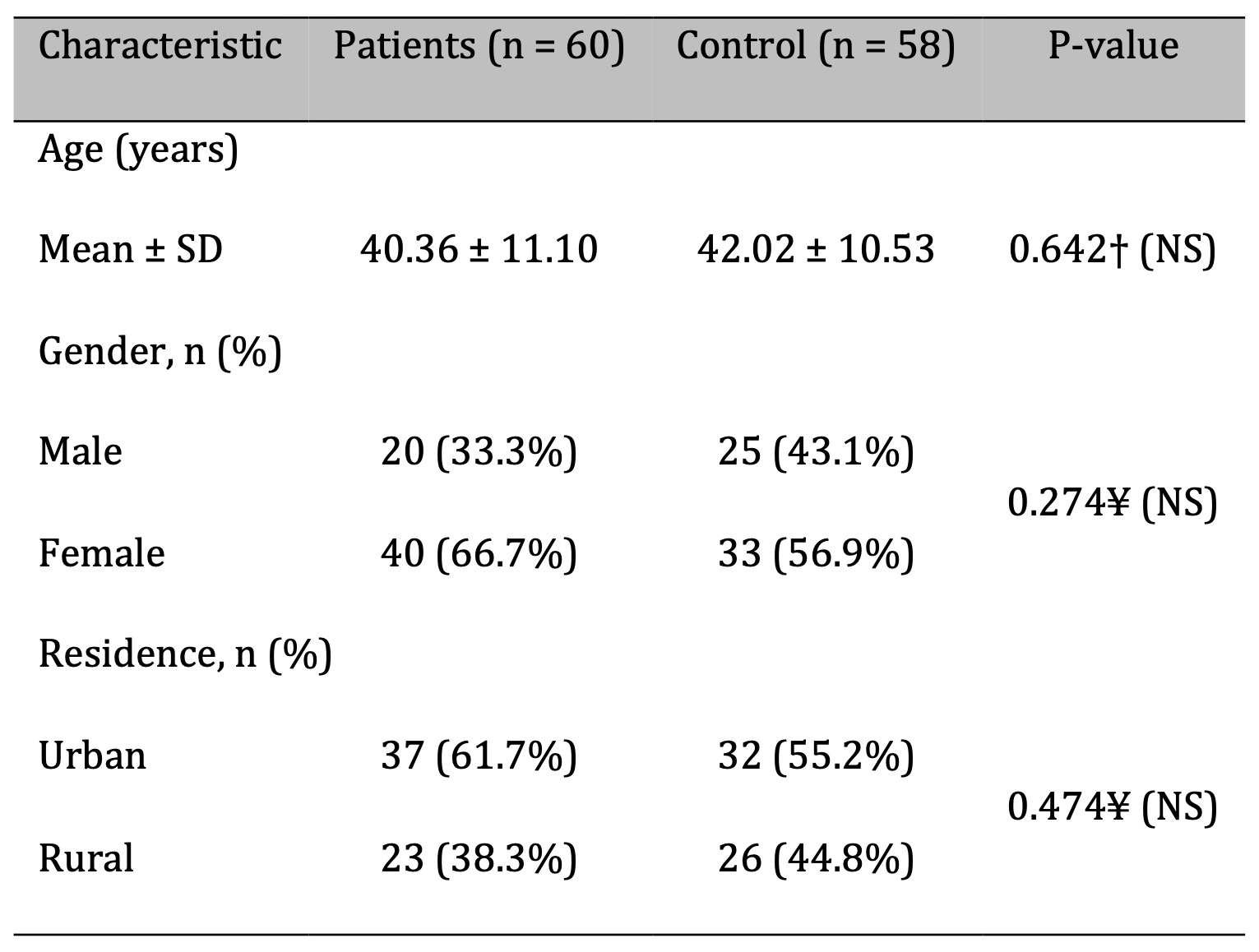
Table 2: Demographic Characteristics of Bronchial Asthmatic Patients and Controls. n: number of cases; SD: standard deviation; †: independent samples t-test; ¥: Chi-square test; NS: not significant at P > 0.05; S: significant at P < 0.05
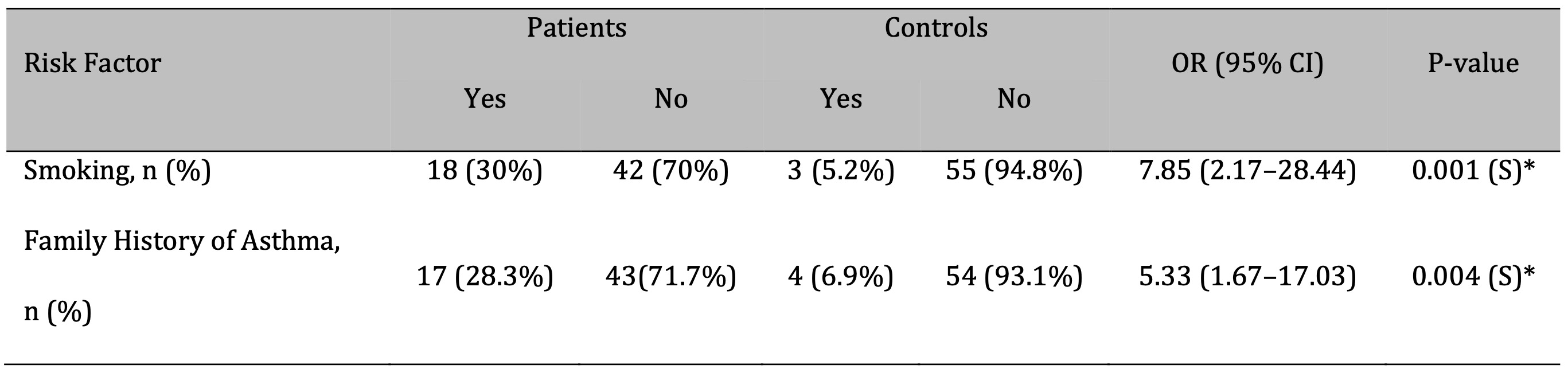
Table 3: Risk Factors Associated with Bronchial Asthma. OR: Odd ratio;CI: Confidence interval
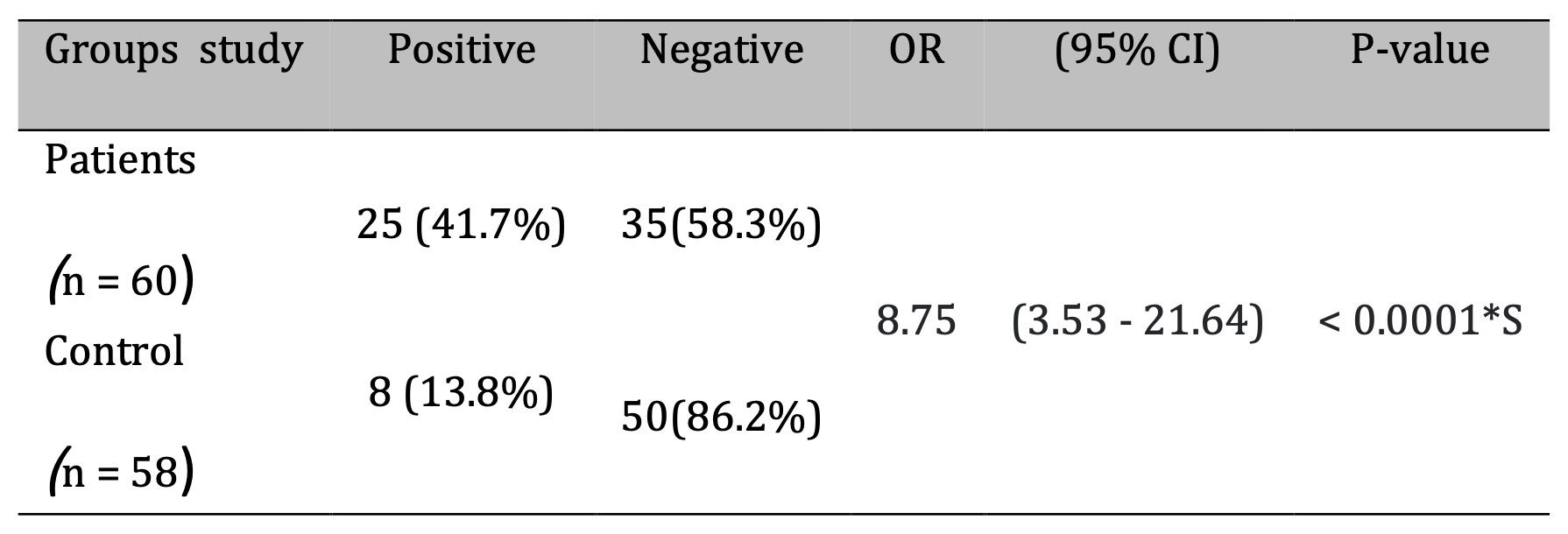
Table 4: Association of Prior M.pneumoniae Infection with Asthma Risk
Association of IL-4 Gene Single-Nucleotide Polymorphism (rs2243250 C>T) with Susceptibility to Bronchial Asthma
Genotypic and allelic frequencies were analyzed to assess the association between the IL-4 rs2243250 polymorphism and susceptibility to bronchial asthma. The genotype distribution among healthy controls was in accordance with Hardy-Weinberg equilibrium (P > 0.05), as presented in Table 5. A significant difference in genotype frequencies was observed between the control group (CC: 68.9%, CT: 24.2%, TT: 6.9%) and the bronchial asthma group (CC: 36.7%, CT: 41.7%, TT: 21.6%) (Figure 2). The T allele was significantly more frequent in asthma patients (42.5%) compared to controls (18.97%) (OR = 3.16, P = 0.0001) (Fig. 3, Table 5). Both the CT (OR: 3.25; 95% CI: 1.41–7.49; P = 0.005) and TT (OR: 5.91; 95% CI: 1.72–20.33; P = 0.004) genotypes were significantly associated with an increased risk of bronchial asthma.
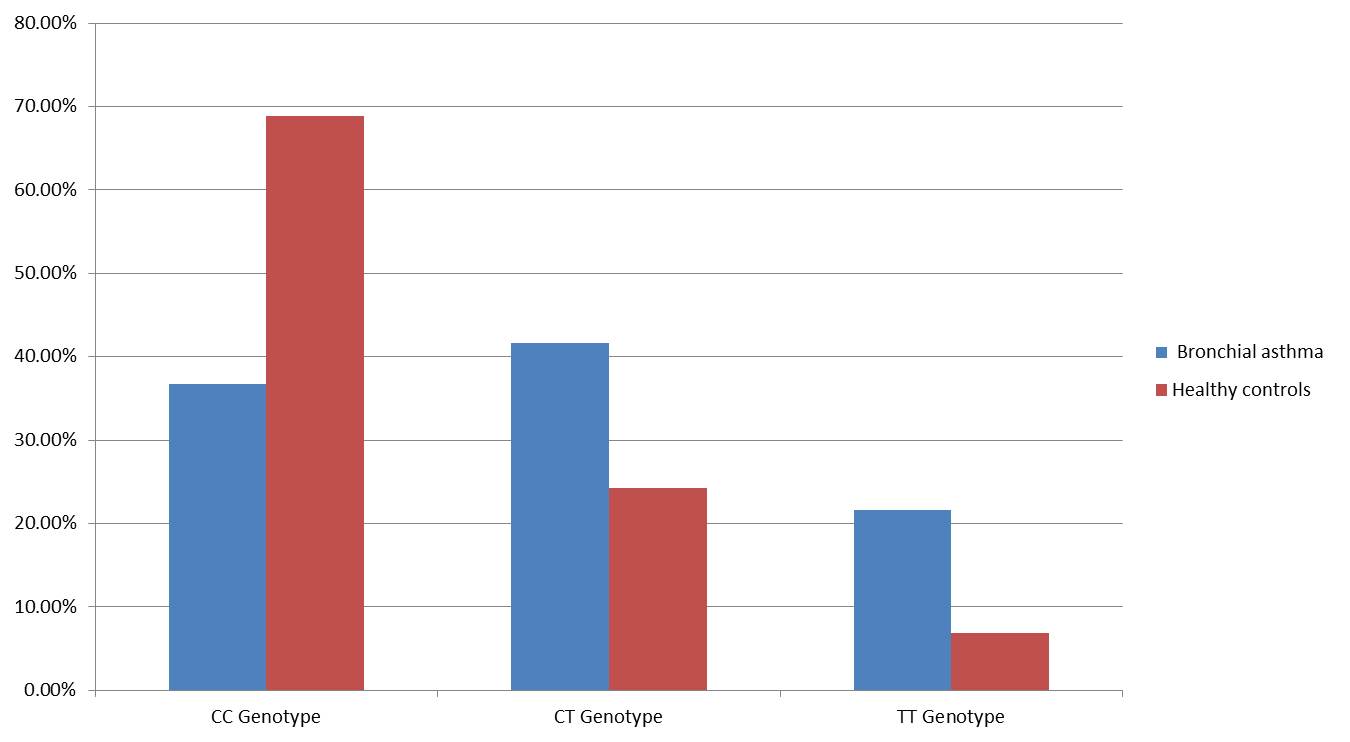
Fig. 2: Genotype frequency among study groups.
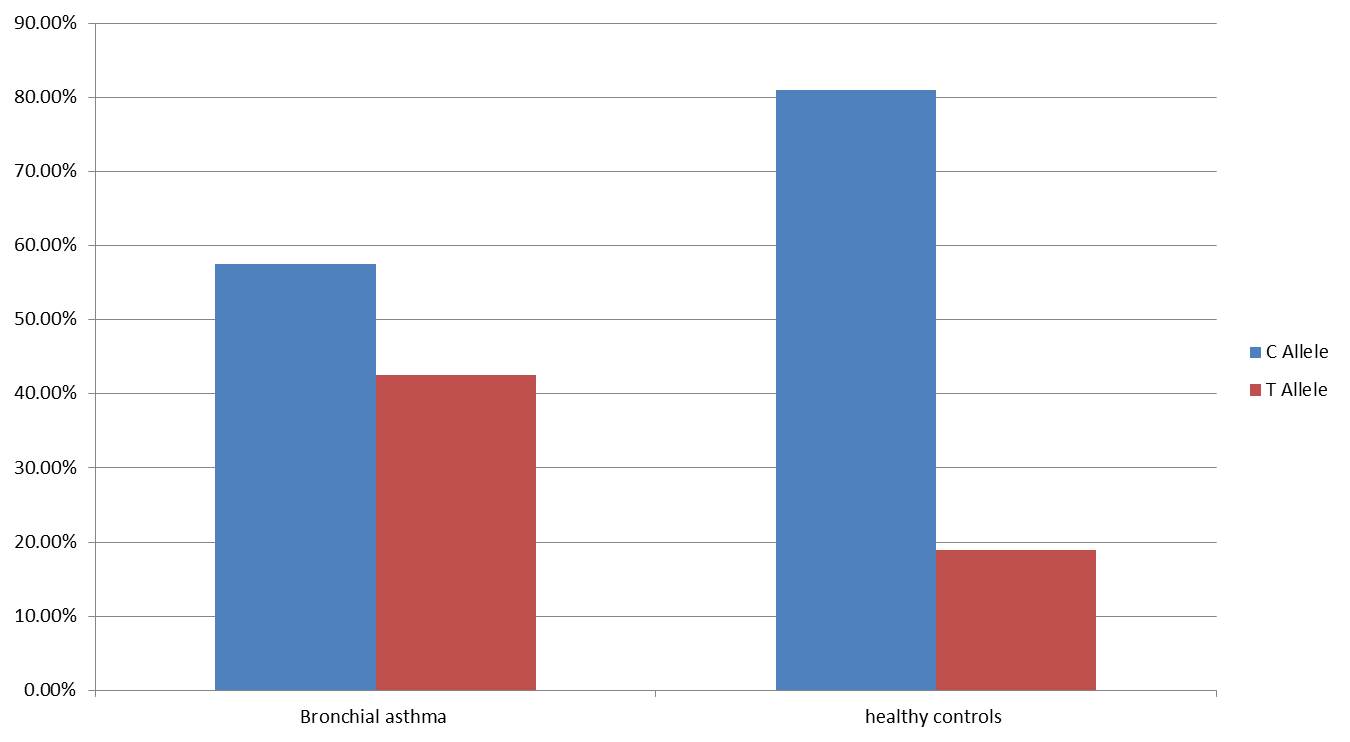
Fig. 3: Wild type and Mutant allele frequency among study groups.
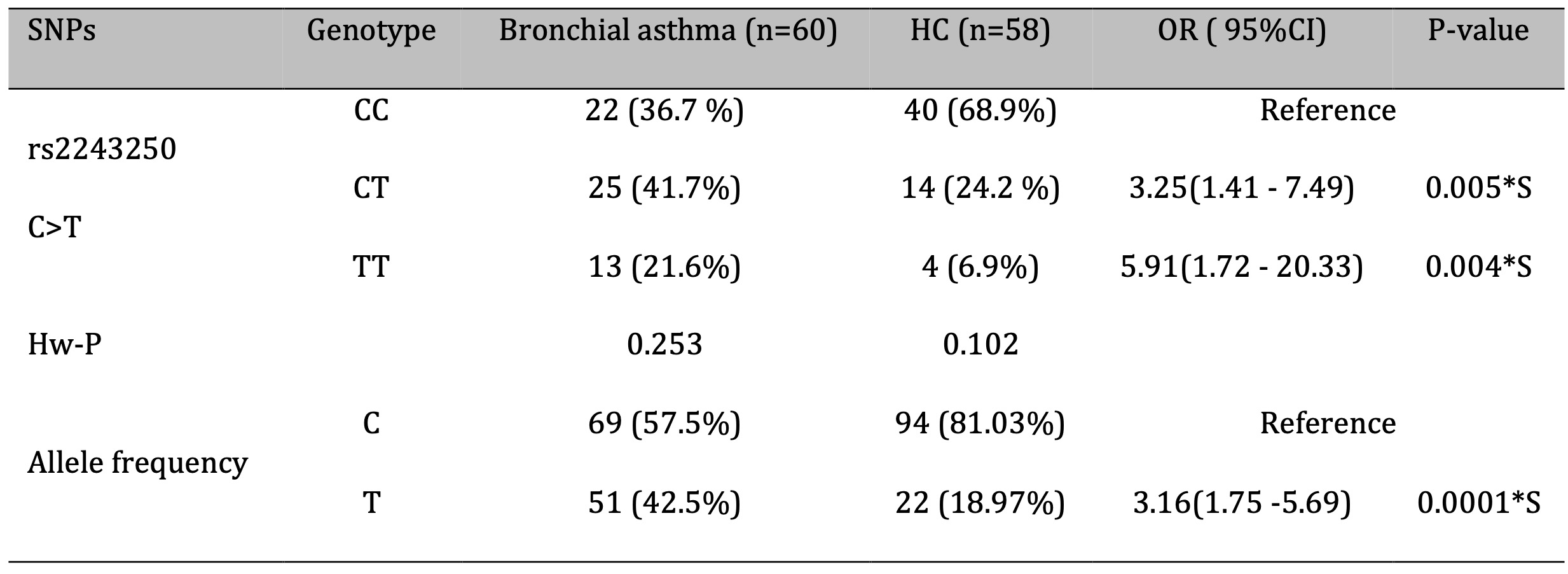
Table 5: Association between IL-4 (rs2243250 C>T) genotypes and bronchial asthma risk and allele frequency distribution. OR: odds ratio; CI: confidence interval; S: significant at P < 0.05
Impact of IL-4 (rs2243250 C>T) Gene Polymorphisms on Serum Levels of IL-4
The association between the IL-4 gene polymorphism—specifically the rs2243250 variant, characterized by a C>T substitution—and serum IL-4 levels in patients with bronchial asthma is presented in Fig. 4. This figure illustrates how this genetic variation may influence IL-4 concentrations in the serum of affected individuals. The analysis demonstrated a statistically significant effect of the IL-4 (rs2243250) polymorphism on serum IL-4 levels (P < 0.001). Patients with the TT genotype showed the highest IL-4 concentrations, significantly greater than those observed in individuals with the TC and CC genotypes (P < 0.001).
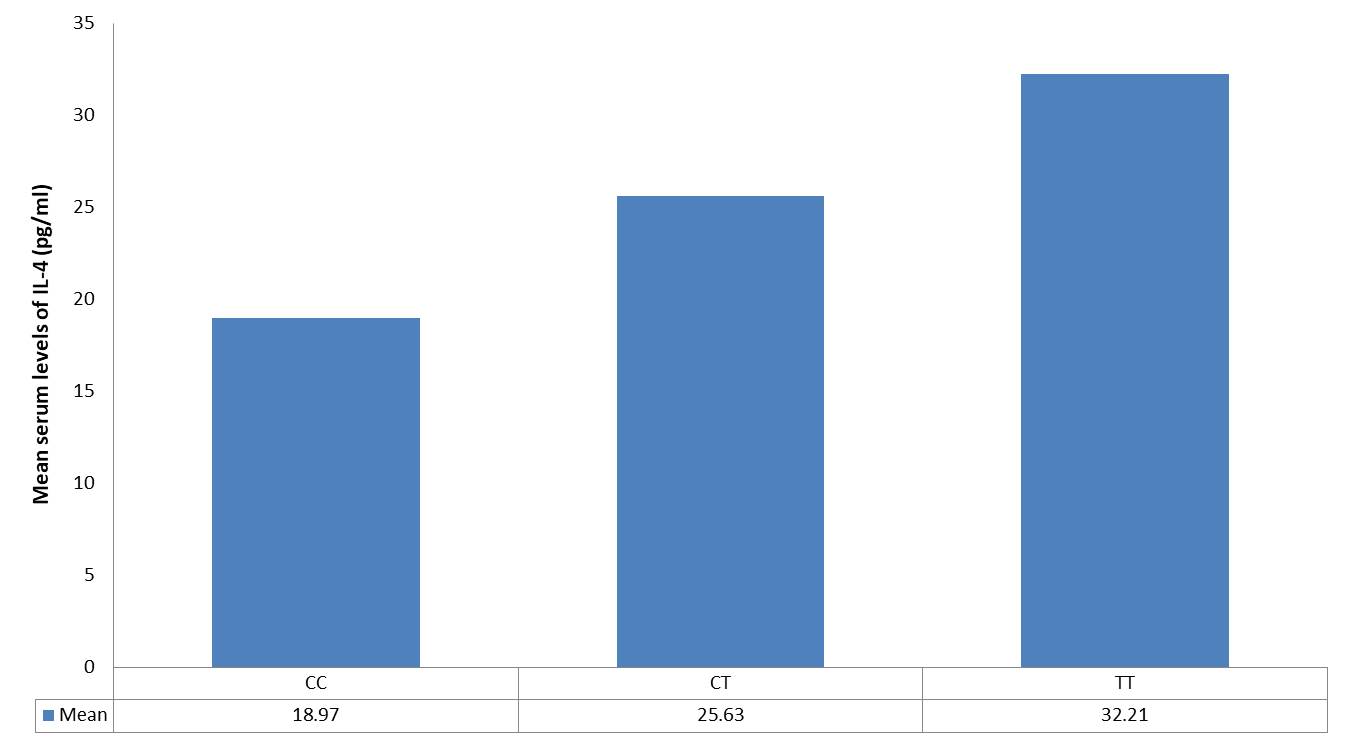
Fig. 4: Association between IL-4 gene polymorphism (rs2243250 C > T) genotypes and serum levels of IL-4 among bronchial asthma patients.
Discussion
This case-control study investigated the association between the IL-4 gene single nucleotide polymorphism (SNP) rs2243250 and susceptibility to bronchial asthma, along with its relationship to serum IL-4 levels in allergic asthma patients. Participants were statistically matched for age, gender, and residence. The asthma patient group included 66.7% females, consistent with findings by [20], who reported a 65% female predominance in asthma cases.
Asthma prevalence and severity are influenced by age and sex. While boys are more commonly affected during childhood, adult women experience higher prevalence and more severe forms of the disease. This disparity is influenced by hormonal changes, genetic predisposition, sociocultural factors, and differential treatment responses. Hormonal fluctuations during puberty, menstruation, and pregnancy are associated with asthma development in women. Additionally, gene-environment interactions contribute to disease outcomes [21].
Smoking was found to significantly increase asthma risk; individuals who smoked were 7.85 times more likely to develop asthma compared to non-smokers. This elevated risk aligns with existing literature, which suggests that tobacco exposure disrupts pulmonary immune responses, promotes Th2-dominated inflammation, and increases infection susceptibility [22, 23]. Even minimal exposure exacerbates asthma symptoms and increases the frequency of attacks [24]. A family history of asthma also increased disease risk by 5.33 times. Asthma’s polygenic nature and strong familial aggregation suggest heritability estimates between 60% and 80% [25].
Patients exhibited a significantly higher prevalence of prior Mycoplasma pneumoniae infection (41.7%) compared to controls (13.8%), suggesting that M. pneumoniae may contribute to increased susceptibility in affected individuals, either as a risk factor or a triggering event (OR = 8.75, p < 0.0001). This observation aligns with earlier epidemiological studies reporting a higher incidence of M. pneumoniae infection among asthmatic individuals compared to non-asthmatic controls [26, 27, 28].
The immunomodulatory properties and associated inflammatory responses elicited by M. pneumoniae may exacerbate or initiate immune dysregulation, contributing to the observed clinical manifestations. Further investigation is warranted into mechanisms such as cytokine dysregulation, molecular mimicry, and persistent infection [29]. M. pneumoniae is frequently associated with asthma exacerbations and is known to increase hospitalization and emergency room visits during outbreaks. Although the precise mechanisms remain unclear, its role in asthma pathogenesis likely involves epithelial damage, mucus hypersecretion, and airway inflammation [30].
Guo et al. (2024) further explored the association between M. pneumoniae and the onset or worsening of asthma, highlighting the interplay between host genetic susceptibility and infectious agents in asthma pathogenesis. Their findings underscore the need to consider both genetic and environmental factors when examining asthma etiology and exacerbation. The high prevalence of M. pneumoniae infection in asthma patients underscores the importance of further research to clarify its role in disease progression and to inform therapeutic or preventive strategies [31].
Genetic polymorphisms in inflammatory mediator genes, particularly cytokines, play a significant role in disease susceptibility [32]. In the present study, a significant difference in the distribution of IL-4 rs2243250 genotypes and alleles was observed between asthma patients and healthy controls. Specifically, the CT (heterozygous) and TT (homozygous mutant) genotypes were significantly associated with an increased risk of bronchial asthma. Individuals carrying the T allele were found to have a 3.16-fold higher likelihood of developing asthma compared to those carrying the C allele.
Several studies have suggested a link between the IL-4 -590 C>T polymorphism and asthma risk. Vercelli (2008) reported a strong association between the T allele and asthma severity in British patients [33], while Neelofar et al. (2017) found that the T allele was associated with asthma and elevated serum IgE levels in children. These findings are consistent with our results, which also demonstrate a significant association between the T allele and asthma [24]. Zhang et al. (2019) similarly reported a correlation between the IL-4 -590 C>T polymorphism and bronchial asthma in Uyghur children, noting increased IgE levels and decreased FEV1, suggesting that the T allele may have a pathogenic role in this population [35].
Despite this evidence, the association between the IL-4 -590 T allele and asthma susceptibility remains debated. Polymorphisms in the promoter region of the IL4 gene have been linked to increased asthma risk, as IL-4 plays a central role in asthma pathophysiology. IL-4 promotes the differentiation of naïve T cells into Th2 cells, inhibits Th1 responses, and stimulates B cell proliferation and IgE production by enhancing the expression of MHC class II, CD40, CD23, and FcεRI. It also promotes endothelial proliferation and upregulates vascular cell adhesion molecule-1 (VCAM-1), facilitating the inflammatory cascade associated with asthma. Notably, several asthma phenotypes are associated with IL4 gene polymorphisms [17].
Analysis of IL4 rs2243250 also revealed a significant influence on serum IL-4 levels. Individuals with the TT genotype exhibited significantly higher cytokine levels compared to those with TC or CC genotypes. Rosenwasser et al. (1995) first implicated the C>T polymorphism at position -590 in the IL4 promoter in asthma development, showing that the T allele was associated with elevated IgE levels. This single nucleotide polymorphism enhances IL4 gene expression by increasing transcription factor binding, contributing to elevated IL-4 production and stronger inflammatory responses [36]. Numerous studies support the role of high IL4-590C/T gene expression in bronchial asthma [37, 38], although some have reported conflicting results [39]. These discrepancies may reflect differences in study design, sample size, population genetics, or geographic factors.
Study Limitations
This study has several limitations. The relatively small sample size and the fact that the study was confined to a single geographic area may limit applicability to other regions or ethnic groups. Potential confounding factors, such as environmental exposures (e.g., air pollutants, allergens), socioeconomic conditions, and other unmeasured genetic variants, may also have influenced the outcomes.
Clinical and Public Health Implications
Despite these limitations, the study highlights important associations between genetic variation, infection history, and asthma risk. Genotyping of IL-4 rs2243250 may aid in identifying individuals at heightened risk of developing asthma and inform targeted therapeutic strategies. From a public health perspective, strategies to prevent M. pneumoniae infection, particularly among genetically susceptible individuals, may reduce asthma onset or severity. These findings support the integration of genetic screening and infection control into comprehensive asthma management and prevention programs.
Conclusion
The IL4 rs2243250 variant is significantly associated with increased asthma susceptibility and elevated serum IL-4 levels in the Iraqi population. However, larger, more diverse studies are needed to validate these findings. Additionally, the strong association observed between prior Mycoplasma pneumoniae infection and asthma highlights its potential clinical significance and justifies further investigation into its role in disease pathogenesis and as a possible target for therapeutic or preventive interventions.
Acknowledgements
The authors sincerely thank all professionals and collaborators whose efforts and support contributed to the successful completion of this research.
Author Contributions
All authors made equal contributions to the study design, data collection, analysis, manuscript preparation, revision, and final approval of the submitted version.
Financial Support
The authors declare that no financial support was received from any organization for this study.
Ethical Approval
This study received ethical approval from the Babylon Health Directorate (Approval No. 68, dated 21/01/2021). Verbal informed consent was obtained from all participants.
Disclosure Statement
The authors declare no conflicts of interest related to this work. The content of this paper has been written without assistance of AI-based services.
References
| 1 | Li HF, Yan LP, Wang K, Li XT, Liu HX, Tan W. Association between ADAM33 polymorphisms and asthma risk: a systematic review and meta-analysis. Respir Res. 2019 Feb 21;20(1):38 doi: 10.1186/s12931-019-1006-1 PMID: 30791911; PMCID: PMC6385425.
https://doi.org/10.1186/s12931-019-1006-1 |
| 2 | Bush A. Pathophysiological Mechanisms of Asthma. Front Pediatr. 2019 Mar 19;7:68 doi: 10.3389/fped.2019.00068 PMID: 30941334; PMCID: PMC6434661.
https://doi.org/10.3389/fped.2019.00068 |
| 3 | Baïz N, Annesi-Maesano I. Is the asthma epidemic still ascending? Clin Chest Med. 2012 Sep;33(3):419-29 doi: 10.1016/j.ccm.2012.06.001 Epub 2012 Jul 18 PMID: 22929092.
https://doi.org/10.1016/j.ccm.2012.06.001 |
| 4 | Bateman ED, Hurd SS, Barnes PJ, Bousquet J, Drazen JM, FitzGerald JM, Gibson P, Ohta K, O'Byrne P, Pedersen SE, Pizzichini E, Sullivan SD, Wenzel SE, Zar HJ. Global strategy for asthma management and prevention: GINA executive summary. Eur Respir J. 2008 Jan;31(1):143-78 doi: 10.1183/09031936.00138707 Erratum in: Eur Respir J. 2018 Jan 31;51(2):0751387 doi: 10.1183/13993003.51387-2007 PMID: 18166595.
https://doi.org/10.1183/13993003.51387-2007 |
| 5 | Harfi H, Al Abbad K, Alsaeed AH. Decreased prevalence of allergic rhinitis, asthma and eczema in Riyadh city, Saudi Arabia. Trends Med Res. 2010;5(2):57-62 doi: 10.3923/tmr.2010.57.62
https://doi.org/10.3923/tmr.2010.57.62 |
| 6 | Ziyab AH, Abul AT. Trends in asthma hospital admissions and mortality in Kuwait, 2000-2014: a national retrospective observational study. BMJ Open. 2018 May 8;8(5):e021244 doi: 10.1136/bmjopen-2017-021244 PMID: 29739784; PMCID: PMC5942427.
https://doi.org/10.1136/bmjopen-2017-021244 |
| 7 | Busse WW, Lemanske RF Jr. Asthma. N Engl J Med. 2001 Feb 1;344(5):350-62 doi: 10.1056/NEJM200102013440507 PMID: 11172168.
https://doi.org/10.1056/NEJM200102013440507 |
| 8 | Boyce JA. Mast cells: beyond IgE. J Allergy Clin Immunol. 2003 Jan;111(1):24-32; quiz 33 doi: 10.1067/mai.2003.60 PMID: 12532090.
https://doi.org/10.1067/mai.2003.60 |
| 9 | Subbarao P, Mandhane PJ, Sears MR. Asthma: epidemiology, etiology and risk factors. CMAJ. 2009 Oct 27;181(9):E181-90 doi: 10.1503/cmaj.080612 Epub 2009 Sep 14 PMID: 19752106; PMCID: PMC2764772.
https://doi.org/10.1503/cmaj.080612 |
| 10 | White GE, Mazurek JM, Storey E. Employed adults with asthma who have frequent workplace exposures. J Asthma. 2015 Feb;52(1):46-51 doi: 10.3109/02770903.2014.944984 Epub 2014 Jul 29 PMID: 25029228; PMCID: PMC4554696.
https://doi.org/10.3109/02770903.2014.944984 |
| 11 | Giavina-Bianchi P, Kalil J. Mycoplasma pneumoniae infection induces asthma onset. J Allergy Clin Immunol. 2016 Apr;137(4):1024-1025 doi: 10.1016/j.jaci.2015.11.011 Epub 2016 Jan 12 PMID: 26792205.
https://doi.org/10.1016/j.jaci.2015.11.011 |
| 12 | Hong SJ. The Role of Mycoplasma pneumoniae Infection in Asthma. Allergy Asthma Immunol Res. 2012 Mar;4(2):59-61 doi: 10.4168/aair.2012.4.2.59 Epub 2012 Feb 21 PMID: 22379599; PMCID: PMC3283794.
https://doi.org/10.4168/aair.2012.4.2.59 |
| 13 | Pelaia C, Heffler E, Crimi C, Maglio A, Vatrella A, Pelaia G, Canonica GW. Interleukins 4 and 13 in Asthma: Key Pathophysiologic Cytokines and Druggable Molecular Targets. Front Pharmacol. 2022 Mar 8;13:851940 doi: 10.3389/fphar.2022.851940 PMID: 35350765; PMCID: PMC8957960.
https://doi.org/10.3389/fphar.2022.851940 |
| 14 | Ranjbar M, Whetstone CE, Omer H, Power L, Cusack RP, Gauvreau GM. The Genetic Factors of the Airway Epithelium Associated with the Pathology of Asthma. Genes (Basel). 2022 Oct 15;13(10):1870 doi: 10.3390/genes13101870 PMID: 36292755; PMCID: PMC9601469.
https://doi.org/10.3390/genes13101870 |
| 15 | Wang ZD, Lian D, Shen JL, Sun R, Xu W, Xin Z, Lei L, Jin LH, Jin SD. Association between the interleukin-4, interleukin-13 polymorphisms and asthma: a meta-analysis. Mol Biol Rep. 2013 Feb;40(2):1365-76 doi: 10.1007/s11033-012-2180-0 Epub 2012 Oct 17 PMID: 23070918.
https://doi.org/10.1007/s11033-012-2180-0 |
| 16 | Jin X, Zheng J. IL-4-C-590T locus polymorphism and susceptibility to asthma in children: a meta-analysis. J Pediatr (Rio J). 2021 May-Jun;97(3):264-272 doi: 10.1016/j.jped.2020.05.005 Epub 2020 Aug 9 Erratum in: J Pediatr (Rio J). 2022 Jan-Feb;98(1):111 doi: 10.1016/j.jped.2021.11.001 PMID: 32781035; PMCID: PMC9432276.
https://doi.org/10.1016/j.jped.2021.11.001 |
| 17 | Zhu L, Liu T, Wang L, Li Q, Wu Y, Liu B. Polymorphisms in the interleukin 4 promoter -589C/T gene and the risk of asthma: a systematic review and meta-analysis. Transl Pediatr. 2021 Sep;10(9):2355-2365 doi: 10.21037/tp-21-419 PMID: 34733676; PMCID: PMC8506068.
https://doi.org/10.21037/tp-21-419 |
| 18 | Alsaid A, El-Missiry M, Hatata el-S, Tarabay M, Settin A. Association of IL-4-590 C>T and IL-13-1112 C>T gene polymorphisms with the susceptibility to type 2 diabetes mellitus. Dis Markers. 2013;35(4):243-7 doi: 10.1155/2013/107470 Epub 2013 Sep 9 PMID: 24167373; PMCID: PMC3782814.
https://doi.org/10.1155/2013/107470 |
| 19 | Howell WM, Turner SJ, Theaker JM, Bateman AC. Cytokine gene single nucleotide polymorphisms and susceptibility to and prognosis in cutaneous malignant melanoma. Eur J Immunogenet. 2003 Dec;30(6):409-14 doi: 10.1111/j.1365-2370.2003.00425.x. PMID: 14675394.
https://doi.org/10.1111/j.1365-2370.2003.00425.x |
| 20 | Dimitrova D, Youroukova V, Ivanova-Todorova E, Tumangelova-Yuzeir K, Velikova T. Serum levels of IL-5, IL-6, IL-8, IL-13 and IL-17A in pre-defined groups of adult patients with moderate and severe bronchial asthma. Respir Med. 2019 Jul-Aug;154:144-154 doi: 10.1016/j.rmed.2019.06.024 Epub 2019 Jun 26 PMID: 31260861.
https://doi.org/10.1016/j.rmed.2019.06.024 |
| 21 | Chowdhury NU, Guntur VP, Newcomb DC, Wechsler ME. Sex and gender in asthma. Eur Respir Rev. 2021 Nov 17;30(162):210067 doi: 10.1183/16000617.0067-2021 PMID: 34789462; PMCID: PMC8783601.
https://doi.org/10.1183/16000617.0067-2021 |
| 22 | Burke H, Leonardi-Bee J, Hashim A, Pine-Abata H, Chen Y, Cook DG, Britton JR, McKeever TM. Prenatal and passive smoke exposure and incidence of asthma and wheeze: systematic review and meta-analysis. Pediatrics. 2012 Apr;129(4):735-44 doi: 10.1542/peds.2011-2196 Epub 2012 Mar 19 PMID: 22430451.
https://doi.org/10.1542/peds.2011-2196 |
| 23 | Gibbs K, Collaco JM, McGrath-Morrow SA. Impact of Tobacco Smoke and Nicotine Exposure on Lung Development. Chest. 2016 Feb;149(2):552-561 doi: 10.1378/chest.15-1858 Epub 2016 Jan 12 PMID: 26502117; PMCID: PMC4944770.
https://doi.org/10.1378/chest.15-1858 |
| 24 | Neophytou AM, Oh SS, White MJ, Mak ACY, Hu D, Huntsman S, Eng C, Serebrisky D, Borrell LN, Farber HJ, Meade K, Davis A, Avila PC, Thyne SM, Rodríguez-Cintrón W, Rodríguez-Santana JR, Kumar R, Brigino-Buenaventura E, Sen S, Lenoir MA, Williams LK, Benowitz NL, Balmes JR, Eisen EA, Burchard EG. Secondhand smoke exposure and asthma outcomes among African-American and Latino children with asthma. Thorax. 2018 Nov;73(11):1041-1048 doi: 10.1136/thoraxjnl-2017-211383 Epub 2018 Jun 13 PMID: 29899038; PMCID: PMC6225993.
https://doi.org/10.1136/thoraxjnl-2017-211383 |
| 25 | Skadhauge LR, Christensen K, Kyvik KO, Sigsgaard T. Genetic and environmental influence on asthma: a population-based study of 11, 688 Danish twin pairs. Eur Respir J. 1999 Jan;13(1):8-14 doi: 10.1183/09031936.99.13100899 PMID: 10836316.
https://doi.org/10.1183/09031936.99.13100899 |
| 26 | Yavlovich A, Tarshis M, Rottem S. Internalization and intracellular survival of Mycoplasma pneumoniae by non-phagocytic cells. FEMS Microbiol Lett. 2004 Apr 15;233(2):241-6 doi: 10.1016/j.femsle.2004.02.016 PMID: 15063492.
https://doi.org/10.1016/j.femsle.2004.02.016 |
| 27 | Esposito S, Blasi F, Arosio C, Fioravanti L, Fagetti L, Droghetti R, Tarsia P, Allegra L, Principi N. Importance of acute Mycoplasma pneumoniae and Chlamydia pneumoniae infections in children with wheezing. Eur Respir J. 2000 Dec;16(6):1142-6 doi: 10.1034/j.1399-3003.2000.16f21.x. PMID: 11292120.
https://doi.org/10.1034/j.1399-3003.2000.16f21.x |
| 28 | Biscardi S, Lorrot M, Marc E, Moulin F, Boutonnat-Faucher B, Heilbronner C, Iniguez JL, Chaussain M, Nicand E, Raymond J, Gendrel D. Mycoplasma pneumoniae and asthma in children. Clin Infect Dis. 2004 May 15;38(10):1341-6 doi: 10.1086/392498 Epub 2004 Apr 29 PMID: 15156467.
https://doi.org/10.1086/392498 |
| 29 | Chu KA, Ou TY, Hung WH, Sung J, Chen W, Lin CL, Hung YM, Wei JC. Mycoplasma pneumonia Infection Is Associated With an Increased Risk of Systemic Lupus Erythematosus: A Nationwide, Retrospective Cohort Study. Front Microbiol. 2022 Apr 21;13:815136 doi: 10.3389/fmicb.2022.815136 PMID: 35531287; PMCID: PMC9069054.
https://doi.org/10.3389/fmicb.2022.815136 |
| 30 | Tsai TA, Tsai CK, Kuo KC, Yu HR. Rational stepwise approach for Mycoplasma pneumoniae pneumonia in children. J Microbiol Immunol Infect. 2021 Aug;54(4):557-565 doi: 10.1016/j.jmii.2020.10.002 Epub 2020 Oct 17 PMID: 33268306.
https://doi.org/10.1016/j.jmii.2020.10.002 |
| 31 | Guo ZQ, Gu SY, Tian ZH, Du BY. A comprehensive review of Mycoplasma pneumoniae infection in chronic lung diseases: recent advances in understanding asthma, COPD, and bronchiectasis. Front Med (Lausanne). 2024 Sep 25;11:1437731 doi: 10.3389/fmed.2024.1437731 Erratum in: Front Med (Lausanne). 2024 Nov 08;11:1512825 doi: 10.3389/fmed.2024.1512825 PMID: 39386750; PMCID: PMC11461384.
https://doi.org/10.3389/fmed.2024.1512825 |
| 32 | Smolnikova MV, Kasparov EW, Malinchik MA, Kopylova KV. Genetic markers of children asthma: predisposition to disease course variants. Vavilovskii Zhurnal Genet Selektsii. 2023 Jul;27(4):393-400 doi: 10.18699/VJGB-23-47 PMID: 37465198; PMCID: PMC10350864.
https://doi.org/10.18699/VJGB-23-47 |
| 33 | Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008 Mar;8(3):169-82 doi: 10.1038/nri2257 PMID: 18301422.
https://doi.org/10.1038/nri2257 |
| 34 | Neelofar K, Ahmad J, Ahmad A, Alam K. Study of IL4-590C/T and IL6-174G/C Gene Polymorphisms in Type 2 Diabetic Patients With Chronic Kidney Disease in North Indian Population. J Cell Biochem. 2017 Jul;118(7):1803-1809 doi: 10.1002/jcb.25853 Epub 2017 Mar 21 PMID: 27996163.
https://doi.org/10.1002/jcb.25853 |
| 35 | Zhang JH, Zhang M, Wang YN, Zhang XY. Correlation between IL-4 and IL-13 gene polymorphisms and asthma in Uygur children in Xinjiang. Exp Ther Med. 2019 Feb;17(2):1374-1382 doi: 10.3892/etm.2018.7096 Epub 2018 Dec 13 PMID: 30680016; PMCID: PMC6327510.
https://doi.org/10.3892/etm.2018.7096 |
| 36 | Rosenwasser LJ, Klemm DJ, Dresback JK, Inamura H, Mascali JJ, Klinnert M, Borish L. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy. 1995 Nov;25 Suppl 2:74-8; discussion 95-6 doi: 10.1111/j.1365-2222.1995.tb00428.x. PMID: 8590350.
https://doi.org/10.1111/j.1365-2222.1995.tb00428.x |
| 37 | Zheng S, Zhu X, Li B, Yang J, Cui Y, Lu G [Correlation of gene polymorphism of interleukin 4 receptor alpha peptide chain and total serum IgE levels in asthmatic children in Guiyang area].. Zhonghua Yi Xue Za Zhi. 2014 Sep 30;94(36):2822-7 Chinese. PMID: 25534099.
|
| 38 | Huang HR, Zhong YQ, Wu JF. The association between IFN-γ and IL-4 genetic polymorphisms and childhood susceptibility to bronchial asthma. Gene. 2012 Feb 15;494(1):96-101 doi: 10.1016/j.gene.2011.09.027 Epub 2011 Nov 28 PMID: 22143036.
https://doi.org/10.1016/j.gene.2011.09.027 |
| 39 | Zhang JH, Zhou GH, Wei TT, Chang ZS. Association between the interleukin 4 gene -590C>T promoter polymorphism and asthma in Xinjiang Uighur children. Genet Mol Res. 2016 Jul 25;15(3). doi: 10.4238/gmr.15038363 PMID: 27525870.
https://doi.org/10.4238/gmr.15038363 |