Corresponding Author: Anna Emilia Matthiessen
Fraunhofer Research Institution for Marine Biotechnology and Cell Technology, University of Lübeck,
Institute for Medical and Marine Biotechnology, Mönkhofer Weg 239a, 23562 Lübeck (Germany)
Tel. +49 451 38444853, Fax +49 451 38444812 , E-Mail anna.matthiessen@uni-luebeck.de
Evaluation of Human Skin-Derived Stem Cell Characteristics After Non-Invasive Quantum Dot Labeling
Heiko Benzina Sandra Schumanna,b Anja Richtera Janina Kiera Charli Krusea,b Anna Emilia Matthiessena,b
aFraunhofer Research Institution for Marine Biotechnology and Cell Technology, Lübeck, Germany, bInstitute for Medical and Marine Biotechnology, University of Lübeck, Lübeck, Germany
Introduction
With recent advances in regenerative medicine and cell-based therapies, there is a strong need for new and improved labeling techniques of cells used either in vitro or in vivo. The main goal is long-term non-invasive imaging and tracking of transplanted cells to monitor their survival, migration, differentiation and regenerative effects [1].
Nanotechnology, especially the use of QDs, offers major advantages for highly sensitive tracking of stem cells. In contrast to organic dyes or fluorescent proteins, QDs are superior due to their photostability and sustained fluorescence intensity [2, 3]. Among various QD types and different coatings [4-6], commercially available QDs for cell labeling discussed here are made from a cadmium selenide core that is surrounded by a zinc sulfide shell. Using a custom targeting peptide CdSe/ZnS nanocrystals can easy be delivered into the cytoplasm of living cells [7, 8].
In a previous study, we reported no adverse effects of CdSe/ZnS QD labeling on the differentiation potential of adult stem cells isolated from rat pancreas [9]. Similar observations for other stem cell types such as embryonic stem cells, induced pluripotent stem cells or mesenchymal ones have recently been published [10-12]. Intriguingly, others have noted changes in the differentiation profile of stem cells after labeling with nanoparticles such as the inhibition of osteogenesis with QD labeled bone marrow mesenchymal stem cells [13-15]. In view of these possible effects of QDs on different cell types, the analysis of an influence for each individual cell type is inevitable.
The core function of human skin adult epidermal stem cells (EpiSCs) is lifelong self-renewal. As a highly proliferative cell population, they are located on the basement membrane between the epidermis and the dermis [16, 17] and pursue their way of terminal differentiation by leaving their niche and migrating vertically to replace damaged or dead cells and thus ensure tissue homeostasis [16, 18].
Because of their promising potential for skin regeneration, EpiSCs have become an attractive tool especially for skin wound healing. The current gold standards of autologous skin grafts or xenogeneic swine-derived grafts are often limited in their graft size combined with functional and aesthetic issues [19]. The aim of matrix-based cell therapies is to ensure complete, scar-free regeneration including skin appendages such as glands and hair follicles, as well as functional vascularization [20, 21]. It is therefore of major importance to closely monitor the application of cells in human wound healing with regard to their distribution, migration, proliferation and differentiation in order to prevent adverse cell functions and minimize the risk for the patients being treated [22].
Looking for a suitable labeling method for further in vivo experiments, we here describe the effects of CdSe/ZnS-QD labeling on human EpiSCs in vitro for the first time in order to rule out a negative effect on the behavior of the cells.
Materials and Methods
Human epidermal stem cells
Human epidermal stem cells (EpiSCs) were isolated according to their ability to adhere to collagen type IV rapidly described by Kim et al. [23].
EpiSCs used in this study were isolated from human abdominal and groin skin biopsies obtained from plastic surgery intervention. Male and female donors were aged between 28 and 58 years to consider patients variability. In brief, the tissue was minced into small pieces and incubated overnight in dispase (2,0 U/ml, Roche, Germany) to detach the epidermis from the dermis. The epidermis was further treated with trypsin (0.05 % in 1x DPBS, PAA, Austria) for 10 min interrupted by resuspension through a 1000 µl pipette tip. The reaction was blocked with 10 % FCS in DPBS (1x, Gibco, Germany) followed by centrifugation for 5 min and 180 g. Cells were seeded on collagen type IV coated culture dishes (BD Biosciences, USA) in EpiLife® defined Growth Medium with EDGS (both Gibco, Germany) and 0.6 % antibiotic-antimycotic mix (100 x, Gibco, Germany). After incubation for 7 min at 37 °C free floating cells were removed, selecting fast adherent cells for further cultivation. Culture medium was changed every 3-4 days and subcultivation took place at 90 % confluency. In this study, cells were analyzed in passage 3.
Quantum dot labeling
Following the manufacturer´s instruction Qdot® Nanocrystals Qtracker® 605 (QD; Molecular Probes®, Germany) were used to label human epidermal stem cells. The core of these nanocrystals is made up of cadmium selenide (CdSe) surrounded by a zinc sulfide (ZnS) shell. The excitation of these nanocrystals ranges from 405 to 565 nm in contrast to their sharp emission at 605 nm. In addition to the recommended concentration (10 nM) a lower (5 nM) and a higher concentration (20 nM) were applied. The labeling kit was composed of nanocrystals and the corresponding carrier, a custom targeting peptide [24, 25], which enabled the uptake by cells. After incubation for 1 h the cells were washed twice with culture medium and were further cultivated before experiments took place.
FACS analysis
To quantify the amount of labeled cells 1 h, 24 h, 48 h and 96 h after QD labeling, fluorescence activated cell sorting (FACS Calibur, BD Biosciences, USA) was performed on EpiSCs labeled with different QD concentrations (5 nM, 10 nM and 20 nM). Therefore, EpiSCs at passage 3 isolated from 3 different donors were seeded in cell culture dishes (diameter: 60 mm) at a density of 8400 cells/cm2 and labeled with QDs 24 h later. For FACS analysis EpiSCs were harvested and resuspended in PBS with 1 % FCS. Furthermore, an unlabeled control was analyzed at every time point.
Time lapse microscopy
EpiSCs at passage 3 from 3 different donors were seeded into 6-well-culture plates at a density of 8800 cells/cm2 and labeled after 24 h of cultivation with 10 nM QDs and 20 nM QDs respectively. Using a time lapse microscope (Axiovert 200M, Zeiss, Germany) combined with an incubator (37°C, 5 % CO2) labeled cells were monitored over a period of 5 days. Images were recorded every 15 min and finally composed to a movie. Unlabeled cells were analyzed as a negative control.
Growth characteristics
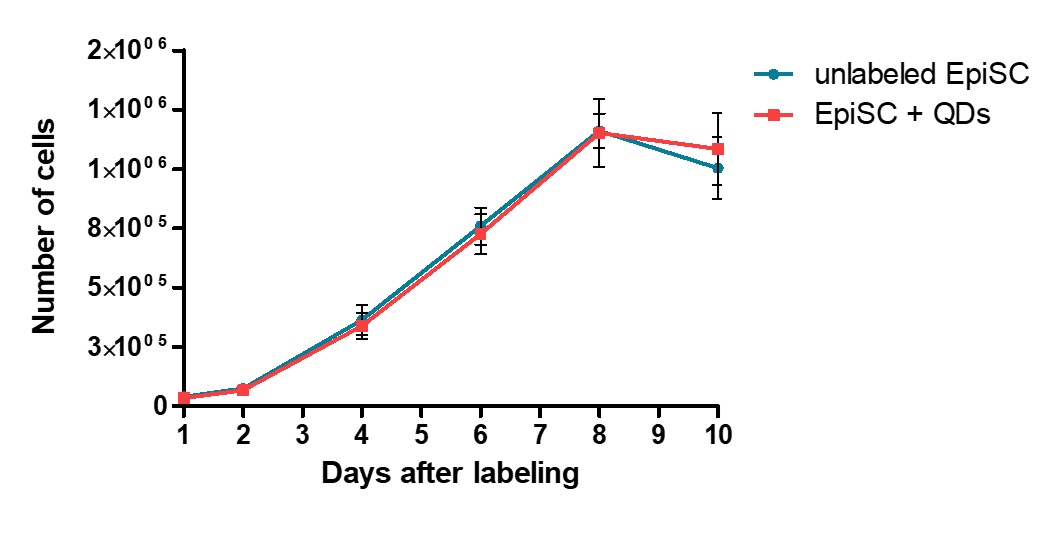
To analyze the impact of QDs on proliferation of human EpiSCs the growth of labeled and unlabeled cells was observed over a period of 10 days. Cells were seeded in triplicates into 6-well culture plates in a density of 4400 cells/cm². One day after seeding, cells were labeled with 10 nM QDs. Unlabeled cells were kept in culture as a control. During the following 10 days the cells were harvested and counted with NucleoCounter (Chemometec, Denmark) at several time points (1, 2, 4, 6, 8 and 10 days). This experiment was done with cells from 3 different patients.
Gene expression analysis
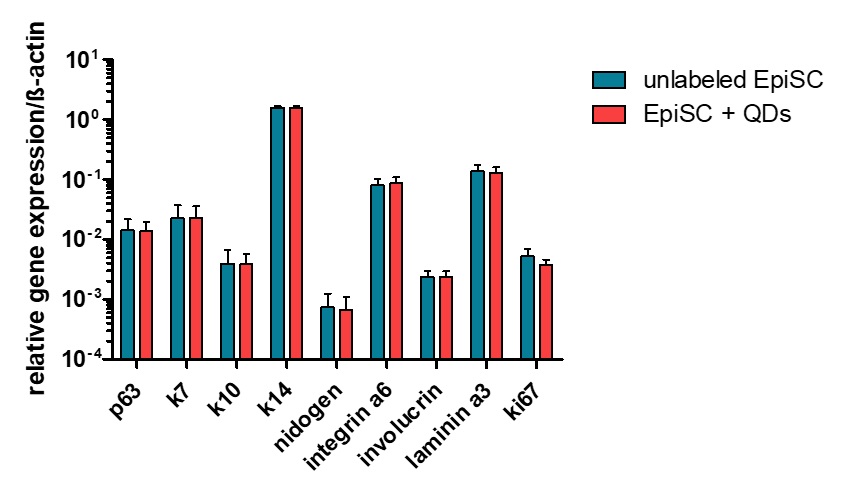
EpiSCs of 3 different donors were seeded in cell culture dishes (diameter: 100 mm) in a density of 1200 cells/cm2 pre-cultured for 5 days. One of each dish was incubated with 10 nM quantum dots and further cultivation of labeled and unlabeled cells took place for 48 h before total RNA was isolated. This was performed using the RNA Plus Mini kit and the QIAcube for automated RNA isolation (both Qiagen, Germany) according to the manufacturer’s protocols. This procedure included a genomic DNA elimination step. RNA concentration was determined by a nanodrop spectrophotometer (PEQLAB Biotechnology, Germany) and cDNA was synthesized from 500 ng template RNA using the QuantiTect reverse transcription kit (Qiagen, Germany) which included a further genomic DNA digestion step. Real-time polymerase chain reaction (PCR) was carried out in with 1 µl cDNA in a 25 µl reaction volume using the QuantiFast SYBR Green PCR kit and human specific QuantiTect primers for: β-actin (149 bp), integrin α6 (142 bp), involucrin (120 bp), keratin 7 (103 bp), keratin 10 (91 bp), keratin 14 (76 bp), ki67 (86 bp), laminin α3 (107 bp), nidogen (123 bp) and p63 (130 bp) (kit and all primer from Qiagen, Germany). The gene expression level of both, labeled and untreated cells, was determined using the housekeeping gene beta-actin as a reference.
Immunocytochemical analysis
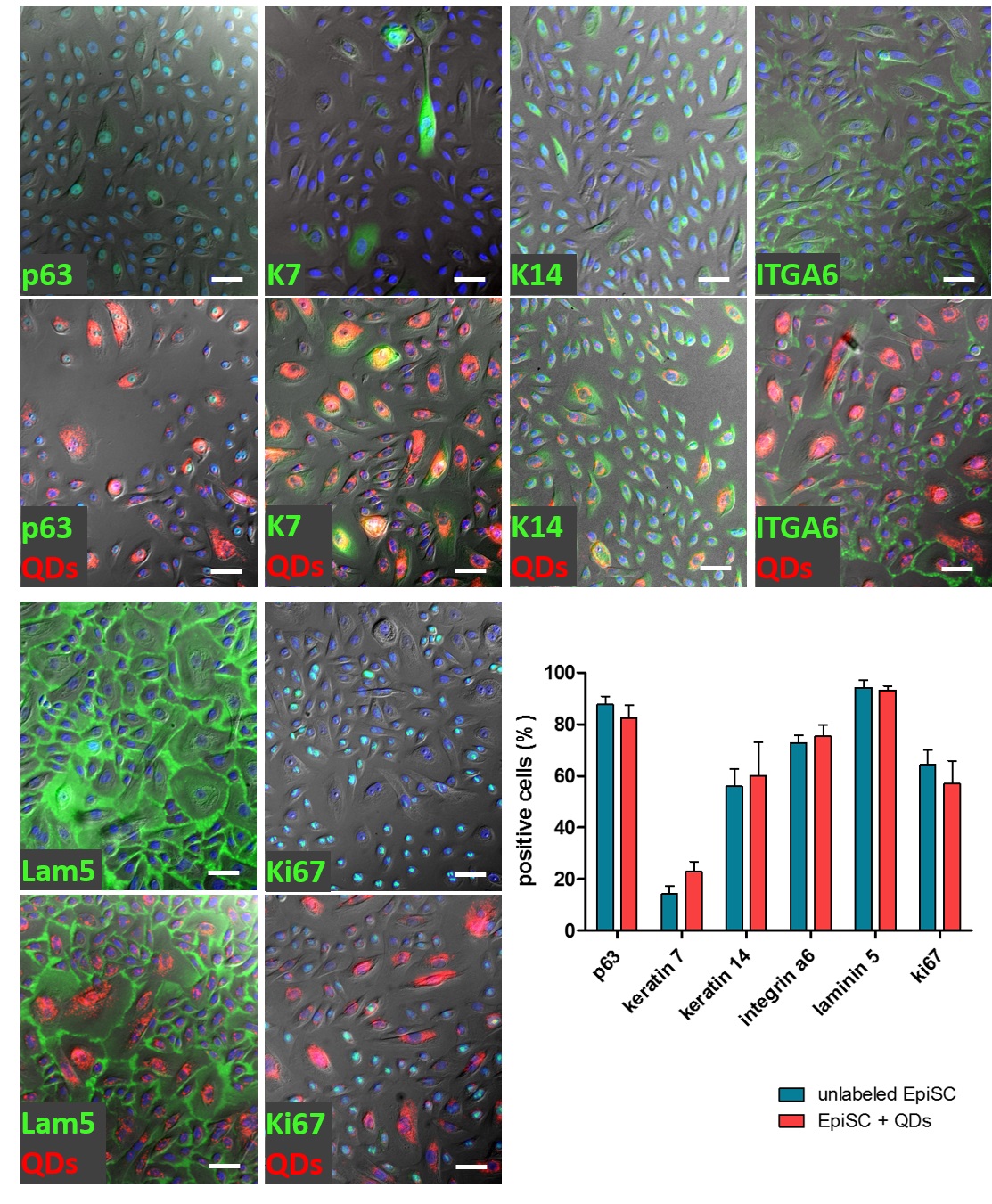
EpiSCs of 3 different donors were seeded on 2-well-chamberslides (3750 cells/cm2; BD Biosciences, USA) and pre-cultured for 5 days. Half of the wells were incubated with 10 nM QDs and further cultivation of labeled and unlabeled cells took place for 48 h. Afterwards the cells were washed with PBS and fixed with 4 % paraformaldehyde (PFA, Merck, Germany) for 15 min at room temperature. Next, cells were rinsed three times with PBS and incubated with 0.1 % Tween 20 (VWR International BDH Prolabo, Germany) for 5 min. Subsequently, samples were blocked with 1.7 % normal goat serum (Vector Laboratories, USA) for the minimum of 20 min at RT. Primary antibodies against: Integrin α6 (1:250, rat monoclonal, Santa Cruz, USA), Keratin 7 (1:500, rabbit polyclonal, Abcam, United Kingdom), Keratin 10 (1:200, mouse monoclonal, Millipore, USA), Keratin 14 (1:500, mouse monoclonal, Santa Cruz, USA), Ki67 (1:500, rabbit polyclonal, Abcam, United Kingdom), Laminin 5 (1:500, mouse monoclonal, Santa Cruz, USA), p63 (1:250, mouse monoclonal, Santa Cruz, USA) diluted in TBS-T (tris-buffered saline-triton X: 150 mM NaCl (Merck, Germany), 10 mM Tris (pH 8.8; biomol, Germany), 0.05 % TritonX (Bio-Rad, USA)) containing 0.1 % bovine serum albumin (PAA Laboratories, Austria) were incubated in a humid chamber for 1h at 37° C. After rinsing three times with PBS samples were incubated under the same conditions with the appropriate secondary antibody: FITC-conjugated anti-rat IgG (1:100), FITC-conjugated anti-rabbit IgG (1:200), FITC-conjugated anti-mouse IgG (1:200, all Jackson ImmunoResearch Europe, United Kingdom) diluted in PBS. Subsequently a nucleus staining with DAPI (1:1000 in PBS; Roche, Switzerland) took place for 5 min followed by washing three times in PBS. Finally, the samples were mounted in Vectashield® mounting medium (Vector Laboratories, USA) and analyzed by fluorescence microscopy Axio Observer Z.1 (Zeiss, Germany). Quantification of marker expression was done by counting representative regions of interest containing from 1900 up to 4400 cells per marker and thereby identifying positive cells.
Results
QD labeling of epidermal stem cells
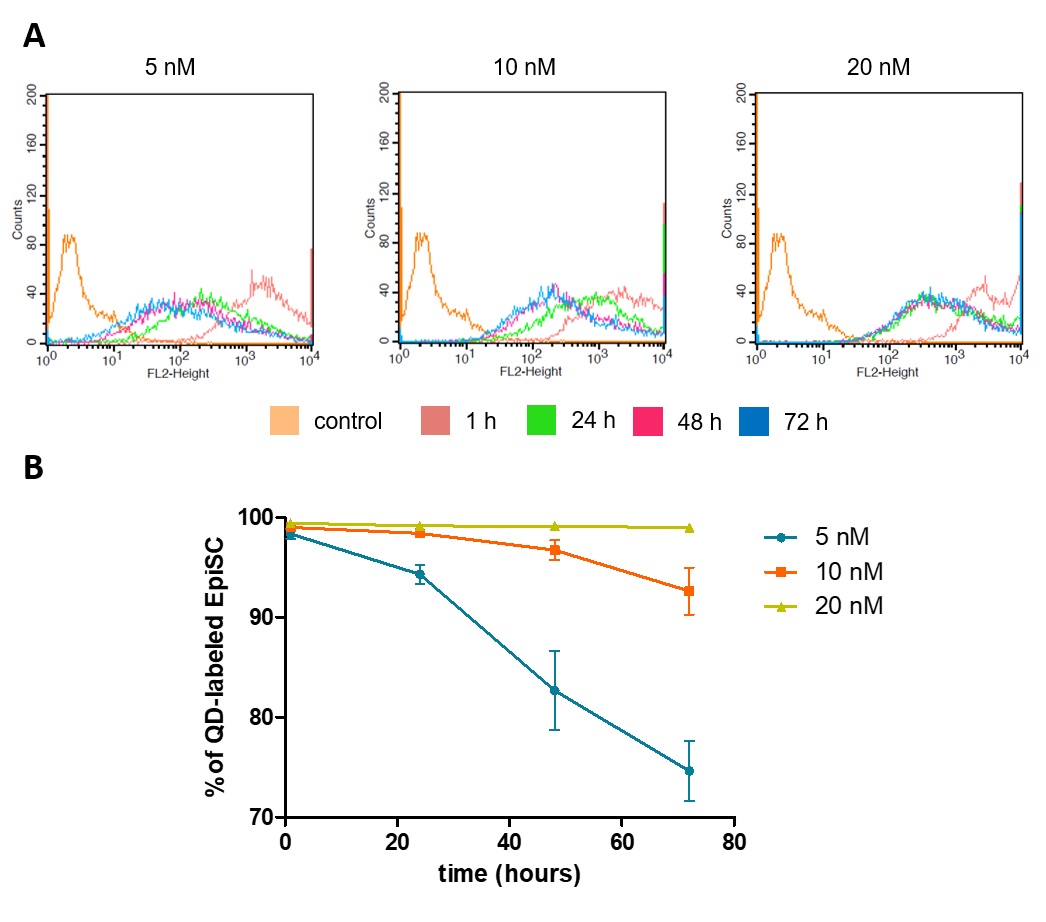
In addition to the manufacturer´s recommendation of 10 nM, a lower (5 nM) and a higher concentration (20 nM) were applied to the cells in order to determine the appropriate QD concentration for labeling EpiSCs. Microscopic analysis of EpiSCs showed a weak signal using 5 nM QDs, while 10 nM and 20 nM QDs resulted in strong fluorescent labeling of the cells with no distinguishable difference between the two concentrations (Fig. 1). FACS analysis, which allowed to monitor the fluorescence signal per cell, provided a deeper insight into QD loading of the cells. Thereby, the highest initial signal could be shown in cells labeled with 20 nM QDs. Furthermore, in the 20 nM approach, the fluorescence signal decreased only within the first 24 h of cultivation and remained stable thereafter, while in the 5 nM and the 10 nM approaches the signal decreased continuously within the monitored period of 72 h (Fig. 2A). In addition to the analysis of the fluorescence intensity, positively labeled cells were quantified over a cultivation period of 72 h. The use of 20 nM QDs resulted in an almost unchanged number of positive cells over the entire period (99.0 %; Fig. 2B), while a slight decrease could be observed using 10 nM QDs (92.6 %) and a more pronounced decrease using 5 nM QDs (74.6 %).
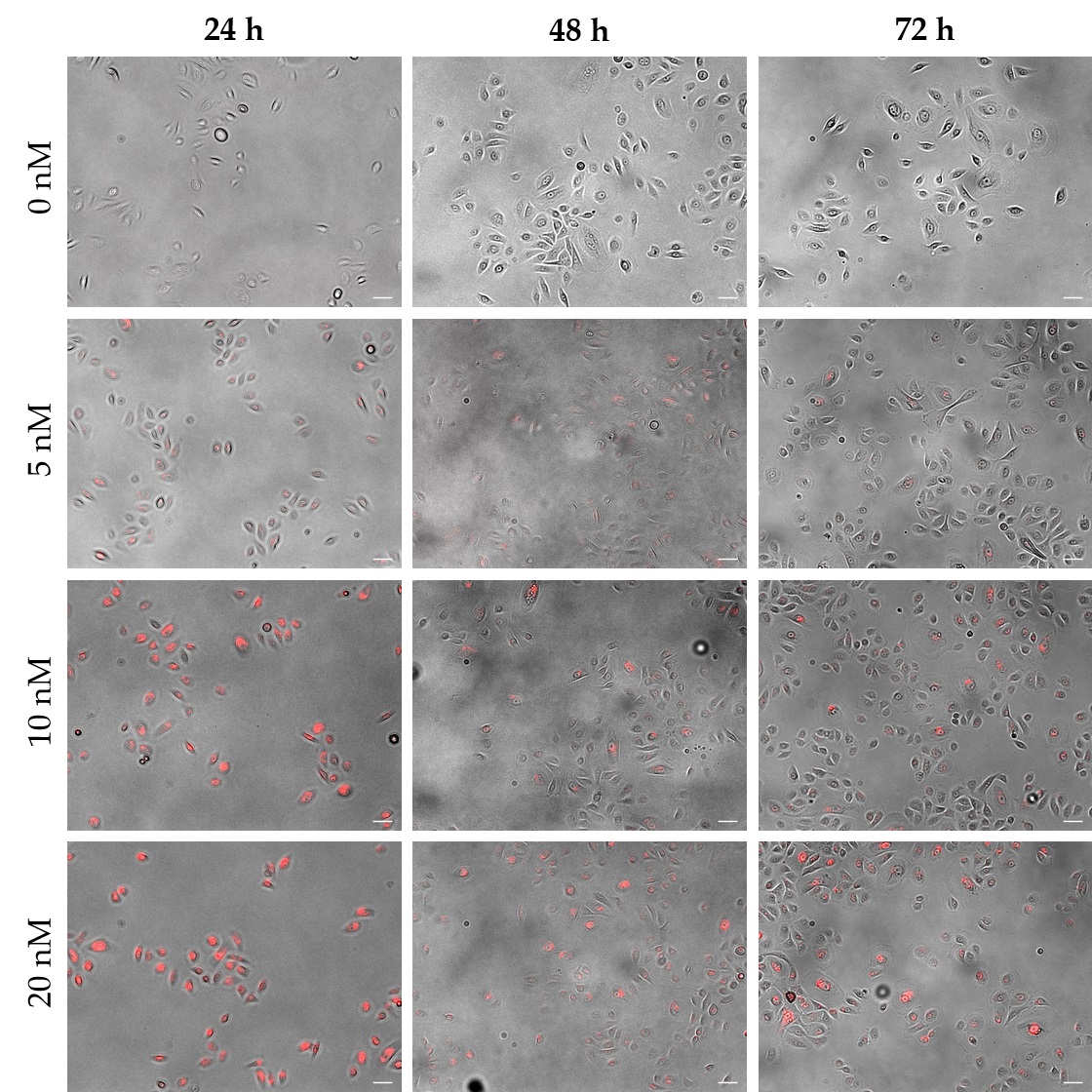
Time lapse analysis of QD distribution during cell division
To investigate how QDs were passed on to daughter cells during cell division, proliferating EpiSCs were imaged after labeling with QDs over a period of 4 days by time lapse microscopy (Fig. 3). Thereby, a thinning of the labeling could be observed. Obviously QD transfer took place in two different ways whereby the distribution of nanoparticles during cell division had an impact on the labeling of daughter cells (Fig. 4). An unsymmetrical distribution of nanoparticles within the cell led to an uneven transmission of QDs after cell division. This resulted in a daughter cell with a stable fluorescent signal on the one hand and a completely unlabeled daughter cell on the other hand. Following a homogeneous distribution of nanoparticles in the cell an equal spread of QDs during cell division was detectable, which led to two daughter cells with a similar amount of QDs. This uniform distribution was always accompanied by a thinning of the QD concentration leading to a constant fading of the fluorescence signal with each cell division.
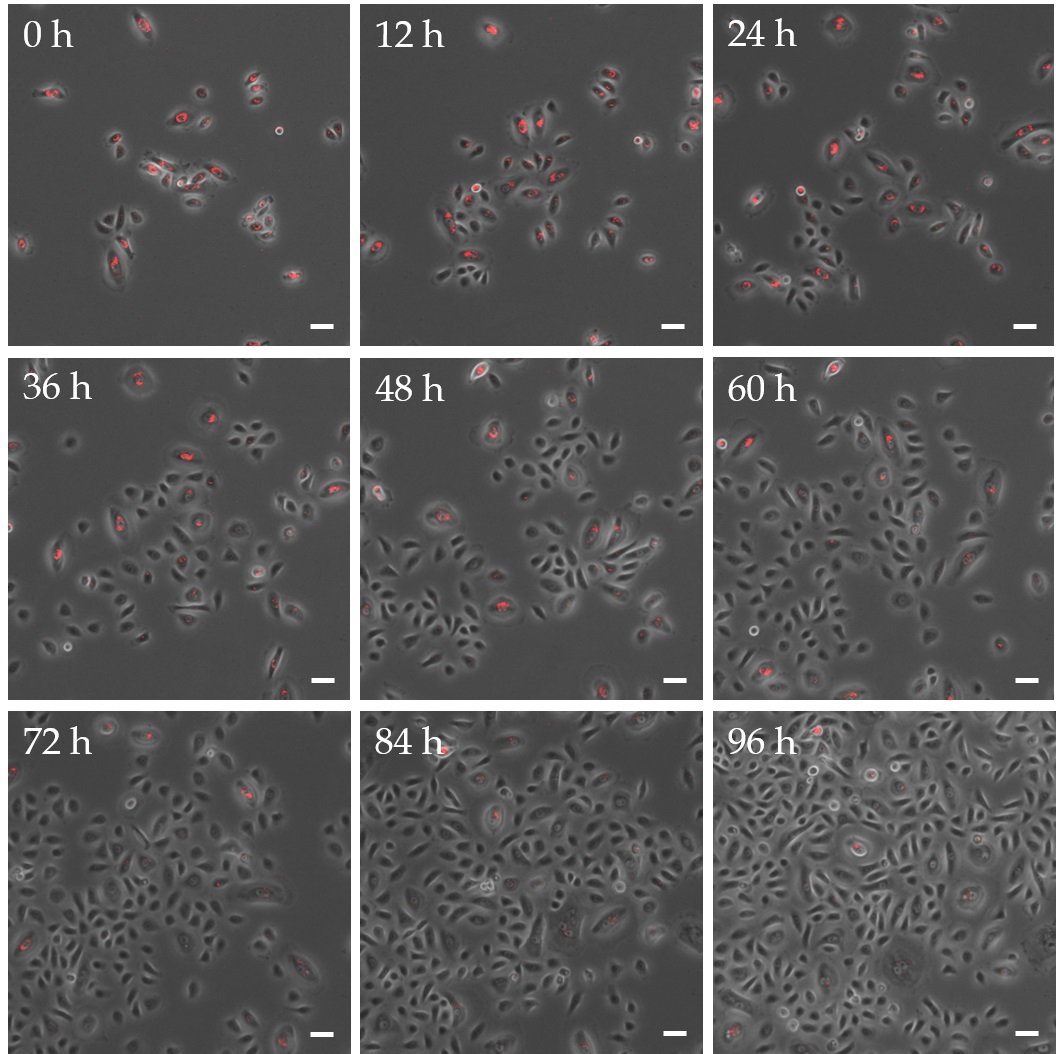
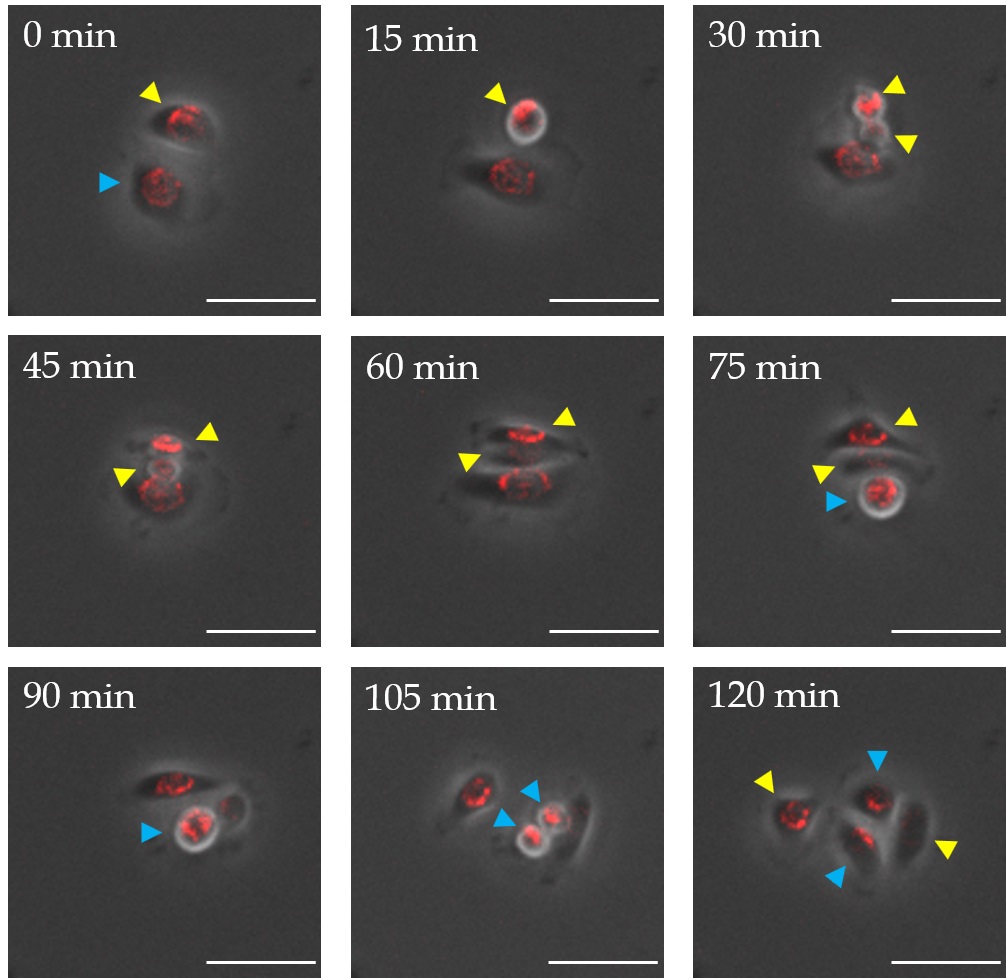
Proliferation characteristics of QD labeled EpiSCs
In order to study the influence of QDs on the proliferation and their long-term labeling potential of human epidermal stem cells in vitro, a growth curve with cells from three patients each was recorded over a period of 10 days with QD labeled and unlabeled cells (Fig. 5). In both conditions a lag phase was followed by a log phase, which then resulted in a stationary phase. No difference in the proliferation capability could be observed, when a labeling concentration of 10 nM was used.
Impact of QDs on the expression of EpiSCs characteristic proteins
Human epidermal stem cells exhibit a very distinct protein expression profile in vitro. In order to rule out adverse effects of QD labeling on epidermal stem cell characteristics, the expression of specific marker proteins was investigated at transcriptional and translational level, performing qRT-PCR and immunocytochemical staining.
The expression level of analyzed genes remained the same after QD labeling of EpiSCs (Fig. 6). Transcripts for the epidermal stem cell marker p63 as well as for epidermal specific keratins like K7, K10, K14 and specific extracellular proteins like integrin α6, involucrin and laminin 3 were still at the same level after labeling with QDs and cultivation for the period of 48 h.
Quantification of immunocytochemical staining revealed also a statistically unchanged amount of marker expressing cells in QD labeled cultures in comparison to unlabeled ones (Fig. 7). All proteins analyzed were still synthesized after QD labeling.
To sum up the results of our study we have demonstrated that human epidermal stem cells could be efficiently labeled with quantum dots. Labeling with QDs had neither an influence on the cell proliferation nor on the characteristics of EpiSCs. From a technical point of view the application of QDs has several advantages. Primarily, the labeling procedure is simple and reproducible. By using a fluorescent microscope no other advanced detection equipment is needed for the further analysis of labeled cells in vitro. Furthermore, the observation of QD cell labeling down to single cell level provides a very good resolution.
The uneven transmission of QDs observed using time-lapse microscopy needs further investigation. One focus could be on the interesting question whether this unequal distribution of QDs is related to the directed distribution of cell organelles and vesicles during asymmetric cell division of epithelial stem cells. If so, that would be an exciting feature of QDs with regard to basic scientific questions on cell division of epithelial stem cells.
Conclusion
Quantum dots are well suited for future tracking experiments in skin organ cultures or even in animal experiments. Even if the particles thin out slightly due to cell proliferation this should not be an obstacle for such ex vivo or in vivo experiments. It is likely that cells transplanted into a tissue will no longer divide so fast but rather differentiate due to the surrounding signals present in the tissue. Therefore, the QD label should be detectable in the cytoplasm of the cells over a longer period of time.
In the following, our cell based in vitro evaluation of quantum dots allows for the application of labeled human epidermal stem cells in a full-thickness human skin model ex vivo. The successful tracing and monitoring of grafted cells and their differentiation potential in this model may pave the way for using quantum dots as a successful, non-invasive and efficient labeling of epidermal stem cells for in vivo applications in skin regeneration and wound healing models.
Funding
This work was founded by the European Regional Development Fund (ERDF).
Statement of Ethics
All experiments were performed in accordance with the World Medical Association Declaration of Helsinki. Utilization of human biopsies for research purposes was approved by the ethics committee of the University of Lübeck (reference number: 10-058). Involved patients gave written informed consent.
The authors have no conflicts of interest to declare.
| 1 Villa C, Erratico S, Razini P, Fiori F, Rustichelli F, Torrente Y, Belicchi M: Stem cell tracking by nanotechnologies. Int J Mol Sci 2010;11:1070-1081. https://doi.org/10.3390/ijms11031070 |
||||
| 2 Alivisatos P: The use of nanocrystals in biological detection. Nat Biotechnol 2004;22:47-52. https://doi.org/10.1038/nbt927 |
||||
| 3 Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S: Quantum dots for live cells, in vivo imaging, and diagnostics. Science 2005;307:538-544. https://doi.org/10.1126/science.1104274 |
||||
| 4 Jaiswal JK, Goldman ER, Mattoussi H, Simon SM: Use of quantum dots for live cell imaging. Nat Methods 2004;1:73-78. https://doi.org/10.1038/nmeth1004-73 |
||||
| 5 Das P, Ganguly S, Banerjee S, Das NC: Graphene based emergent nanolights: a short review on the synthesis, properties and application. Intermed 2019;45:3823-3853. https://doi.org/10.1007/s11164-019-03823-2 |
||||
| 6 Han R, Yu M, Zheng Q, Wang L, Hong Y, Sha Y: A facile synthesis of small-sized, highly photoluminescent, and monodisperse CdSeS QD/SiO(2) for live cell imaging. Langmuir 2009;25:12250-12255. https://doi.org/10.1021/la9016596 |
||||
| 7 Futaki S, Goto S, Suzuki T, Nakase I, Sugiura Y: Structural variety of membrane permeable peptides. Curr Protein Pept Sci 2003;4:87-96. https://doi.org/10.2174/1389203033487261 |
||||
| 8 Umezawa N, Gelman MA, Haigis MC, Raines RT, Gellman SH: Translocation of a beta-peptide across cell membranes. J Am Chem Soc 2002;124:368-369. https://doi.org/10.1021/ja017283v |
||||
| 9 Danner S, Benzin H, Vollbrandt T, Oder J, Richter A, Kruse C: Quantum dots do not alter the differentiation potential of pancreatic stem cells and are distributed randomly among daughter cells. Int J Cell Biol 2013;2013:918242. https://doi.org/10.1155/2013/918242 |
||||
| 10 Liu H, Tang W, Li C, Lv P, Wang Z, Liu Y, Zhang C, Bao Y, Chen H, Meng X, Song Y, Xia X, Pan F, Cui D, Shi Y: CdSe/ZnS Quantum Dots-Labeled Mesenchymal Stem Cells for Targeted Fluorescence Imaging of Pancreas Tissues and Therapy of Type 1 Diabetic Rats. Nanoscale Res Lett 2015;10:959. https://doi.org/10.1186/s11671-015-0959-3 |
||||
| 11 Choi SW, Cho Y, Kim JG, Kim Y, Kim E, Chung H, Kang S: Effect of Cell Labeling on the Function of Human Pluripotent Stem Cell-Derived Cardiomyocytes. Int J Stem Cells 2020;13:287-294. https://doi.org/10.15283/ijsc19138 |
||||
| 12 Grady ST, Britton L, Hinrichs K, Nixon AJ, Watts AE: Persistence of fluorescent nanoparticle‐labelled bone marrow mesenchymal stem cells in vitro and after intra‐articular injection. J Tissue Eng Regen Med 2019;13:191-202. https://doi.org/10.1002/term.2781 |
||||
| 13 Kuo TR, Lee CF, Lin SJ, Dong CY, Chen CC, Tan HY: Studies of intracorneal distribution and cytotoxicity of quantum dots: risk assessment of eye exposure. Chem Res Toxicol 2011;24:253-261. https://doi.org/10.1021/tx100376n |
||||
| 14 Hsieh SC, Wang FF, Hung SC, Chen YJ, Wang YJ: The internalized CdSe/ZnS quantum dots impair the chondrogenesis of bone marrow mesenchymal stem cells. J Biomed Mater Res B Appl Biomater 2006;79:95-101. https://doi.org/10.1002/jbm.b.30517 |
||||
| 15 Hsieh, SC, Wang FF, Lin CS, Chen YJ, Hung SC, Wang YJ: The inhibition of osteogenesis with human bone marrow mesenchymal stem cells by CdSe/ZnS quantum dot labels. Biomaterials 2006;27:1656-1664. https://doi.org/10.1016/j.biomaterials.2005.09.004 |
||||
| 16 Dekoninck S, Blanpain C: Stem cell dynamics, migration and plasticity during wound healing. Nat Cell Biol 2019;21:18-24. https://doi.org/10.1038/s41556-018-0237-6 |
||||
| 17 Rzepka K, Schaarschmidt G, Nagler M, Wohlrab J: Epidermal stem cells. J Dtsch Dermatol Ges 2005;3:962-973. https://doi.org/10.1111/j.1610-0387.2005.05071.x |
||||
| 18 Blanpain C: Stem cells: Skin regeneration and repair. Nature 2010;464:686-687. https://doi.org/10.1038/464686a |
||||
| 19 Schiestl C, Biedermann T, Braziulis E, Hartmann-Fritsch F, Böttcher-Haberzeth S, Arras M, Cesarovic N, Nicolls F, Linti C, Reichmann E, Meuli M: Skingineering II: transplantation of large-scale laboratory-grown skin analogues in a new pig model. Pediatr Surg Int 2011;27:249-254. https://doi.org/10.1007/s00383-010-2792-1 |
||||
| 20 Pang C, Ibrahim A, Bulstrode NW, Ferretti P: An overview of the therapeutic potential of regenerative medicine in cutaneous wound healing. Int Wound J 2017;14:450-459. https://doi.org/10.1111/iwj.12735 |
||||
| 21 Boyce ST, Lalley AL. Tissue engineering of skin and regenerative medicine for wound care. Burns Trauma. 2018;6:4. https://doi.org/10.1186/s41038-017-0103-y |
||||
| 22 Accomasso L, Gallina C, Turinetto V, Giachino C: Stem Cell Tracking with Nanoparticles for Regenerative Medicine Purposes: An Overview. Stem Cells Int 2016;2016:7920358. https://doi.org/10.1155/2016/7920358 |
||||
| 23 Kim DS, Cho HJ, Choi HR, Kwon SB, Park KC: Isolation of human epidermal stem cells by adherence and the reconstruction of skin equivalents. Cell Mol Life Sci 2004;61:2774-2781. https://doi.org/10.1007/s00018-004-4288-4 |
||||
| 24 Futaki S, Goto S, Suzuki T, Nakase I, Sugiura Y: Structural variety of membrane permeable peptides. Curr Protein Pept Sci 2003;4:87-96. https://doi.org/10.2174/1389203033487261 |
||||
| 25 Umezawa N, Gelman MA, Haigis MC, Raines RT, Gellman SH: Translocation of a beta-peptide across cell membranes. J Am Chem Soc 2002 Jan 23;124:368-369. https://doi.org/10.1021/ja017283v |
||||
| 26 Lin S, Xie X, Patel MR, Yang YH, Li Z, Cao F, Gheysens O, Zhang Y, Gambhir SS, Rao JH, Wu JC: Quantum dot imaging for embryonic stem cells. BMC Biotechnol 2007;7:67. https://doi.org/10.1186/1472-6750-7-67 |
||||
| 27 Ryman-Rasmussen JP, Riviere JE, Monteiro-Riviere NA: Surface coatings determine cytotoxicity and irritation potential of quantum dot nanoparticles in epidermal keratinocytes. J Invest Dermatol 2007;127:143-153. https://doi.org/10.1038/sj.jid.5700508 |
||||
| 28 Accomasso L, Gallina C, Turinetto V, Giachino C: Stem Cell Tracking with Nanoparticles for Regenerative Medicine Purposes: An Overview. Stem Cells Int 2016;2016:7920358. https://doi.org/10.1155/2016/7920358 |
||||
| 29 Muller-Borer BJ, Collins MC, Gunst PR, Cascio WE, Kypson AP: Quantum dot labeling of mesenchymal stem cells. J Nanobiotechnology 2007;5:9-18. https://doi.org/10.1186/1477-3155-5-9 |
||||
| 30 Seleverstov O, Zabirnyk O, Zscharnack M, Bulavina L, Nowicki M, Heinrich JM, Yezhelyev M, Emmrich F, O'Regan R, Bader A: Quantum dots for human mesenchymal stem cells labeling. A size-dependent autophagy activation. Nano Lett 2006;6:2826-2832. https://doi.org/10.1021/nl0619711 |
||||
| 31 Hinds KA, Hill JM, Shapiro EM, Laukkanen MO, Silva AC, Combs CA, Varney TR, Balaban RS, Koretsky AP, Dunbar CE: Highly efficient endosomal labeling of progenitor and stem cells with large magnetic particles allow magnetic resonance imaging of single cells. Blood 2003;102:867-872. https://doi.org/10.1182/blood-2002-12-3669 |
||||
| 32 Shapiro EM, Skrtic S, Sharer K, Hill JM, Dunbar CE, Koretsky AP: MRI detection of single particles for cellular imaging. Proc Natl Acad Sci U S A 2004;101:10901-10906. https://doi.org/10.1073/pnas.0403918101 |
||||
| 33 Smirnov P, Lavergne E, Gazeau F, Lewin M, Boissonnas A, Doan B, Gillet B, Combadière C, Combadière B, Clément O: In vivo cellular imaging of lymphocyte trafficking by MRI: A tumor model approach to cell‐based anticancer therapy. Magn Reson Med 2006;56:498-508. https://doi.org/10.1002/mrm.20996 |
||||
| 34 Lagerholm BC, Wang M, Ernst LA, Ly DH, Liu H, Bruchez MP, Waggoner AS: Multicolor Coding of Cells with Cationic Peptide Coated Quantum Dots. Nano Lett 2004;4:2019-2022. https://doi.org/10.1021/nl049295v |
||||
| 35 Morrison SJ, Kimble J: Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 2006;441:1068-1074. https://doi.org/10.1038/nature04956 |
||||
| 36 Pi QM, Zhang, WJ, Zhou GD, Liu W, Cao Y: Degradation or excretion of quantum dots in mouse embryonic stem cells. BMC Biotechnol 2010;10:36. https://doi.org/10.1186/1472-6750-10-36 |
||||
| 37 Xu HN, Tang YY, Ouyang XK: Shear-Induced Breakup of Cellulose Nanocrystal Aggregates. Langmuir 2017;33:235-242. https://doi.org/10.1021/acs.langmuir.6b03807 |
||||
| 38 Lee KH: Quantum dots: a quantum jump for molecular imaging? J Nucl Med 2007;48,1408-1410. https://doi.org/10.2967/jnumed.107.042069 |
||||
| 39 Parak WJ, Pellegrino T, Plank C: Labelling of cells with quantum dots. Nanotechnology 2005;16:9-25. https://doi.org/10.1088/0957-4484/16/2/R01 |
||||
| 40 Li HC, Zhou QF, Liu W, Yan B, Zhao Y, Jiang GB: Progress in the toxicological researches for quantum dots. Sci China Ser B-Chem 2008;51:393-400. https://doi.org/10.1007/s11426-008-0057-9 |
||||
| 41 Derfus AM, Chan WCW, Bhatia SN: Probing the cytotoxicity of semiconductor quantum dots. Adv Mater 2004;4:11-18. https://doi.org/10.1021/nl0347334 |
||||
| 42 Candi E, Cipollone R, Rivetti di Val Cervo P, Gonfloni S, Melino G, Knight R: p63 in epithelial development. Cell Mol Life Sci 2008;65:3126-3133. https://doi.org/10.1007/s00018-008-8119-x |
||||
| 43 Schreder A, Pierard GE, Paquet P, Reginster MA, Pierard-Franchimont C, Quatresooz P: Facing towards epidermal stem cells (Review). Int J Mol Med 2010;26:171-174. https://doi.org/10.3892/ijmm_00000449 |
||||
| 44 Melino G, Memmi EM, Pelicci PG, Bernassola F: Maintaining epithelial stemness with p63. Sci Signal 2015;8:re9. https://doi.org/10.1126/scisignal.aaa1033 |
||||
| 45 Weedon D: Weedon's Skin Pathology. Churchill Livingstone 2011, 3rd Edition, pp 248-249. | ||||
| 46 Ballou B, Lagerholm BC, Ernst LA, Bruchez MP, Waggoner AS: Noninvasive imaging of quantum dots in mice. Bioconjug Chem 2004;15:79-86. https://doi.org/10.1021/bc034153y |
||||