Corresponding Author: Patricia Pereira Coltri
Deptartamento de Biologia Celular e do Desenvolvimento, Instituto de Ciências Biomédicas, Universidade de São Paulo, 05508-000 São Paulo (Brazil)
Tel. +55-11-30910980, E-Mail coltri@usp.br
Human Antigen R (HuR) Facilitates miR-19 Synthesis and Affects Cellular Kinetics in Papillary Thyroid Cancer
Guilherme Henrique Gatti da Silva Maria Gabriela Pereira dos Santos Helder Yudi Nagasse Patricia Pereira Coltri
Departamento de Biologia Celular e do Desenvolvimento, Instituto de Ciências Biomédicas, Universidade de São Paulo, São Paulo, Brazil
Introduction
Human genes are composed of exons and intervening sequences called introns removed during nuclear processing steps. Pre-mRNA splicing is an essential step in eukaryotic gene expression and consists of removing introns and joining exons, resulting in mature mRNAs transcripts [1]. Splicing regulation is dependent on transcript sequences and regulatory proteins recruited by the spliceosome. Interaction between regulatory sequences found across the pre-mRNAs and regulatory proteins defines splicing fate. Additionally, mutations in spliceosome components and splice sites are among the most significant causes of cancer development [2]. More than 70% of the miRNAs are transcribed from introns in the human genome [3, 4]. MicroRNAs (miRNAs) are small non-coding RNAs (20–25 nucleotides) that directly affect gene expression through mediating transcript stability in eukaryotes. miRNAs were described as part of the oncogenic suppressor’s network on tumorigenesis [5, 6]. The oncogenic miRNA cluster miRNA-17-92 is transcribed from intron 3 of MIR17HG gene, located on chromosome 13q31. Seven miRNAs are transcribed as a polycistron, miR17-5p, miR17-3p, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92a-1. Changes in these miRNAs’ expression patterns have been associated with the development of leukemia, ovarian, lung, and thyroid cancer [7, 8].
The miRNAs transcribed from the miR-17-92 cluster miRNAs can perform distinct functions. For example, miR-19a and miR-19b are responsible for the oncogenic and anti-apoptotic effects observed in many lymphomas [8]. Despite being transcribed as a single polycistron, these miRNAs’ processing, maturation, and function are independent [9, 10]. Additionally, individual miRNAs might have variable oncogenic features in different cell types [11, 12].
In papillary thyroid cancer, activation of the mutated BRAFV600E oncogene results in the deregulation of miR-17-92 expression [7]. Among the different targets predicted for miR-17-92 cluster are the receiver (TGFBR1) and the transducer mRNAs (SMAD2, SMAD3 and SMAD4) present in the TGFβ signaling pathway, an essential track in thyroid cell mitogen activation [13-15]. Regulation of transcription of this cluster might also be related to c-MYC and E2F factors [16]. Indeed, the silencing of Pim-1 and E2F3, controlled by c-MYC and E2F, respectively, can regulate the transcription of this cluster [16, 17]. The relative expression of individual miRNAs from this cluster is important for disease development, yet the mechanism that controls miR-17-92 biogenesis remains unknown. We previously found Human antigen R (HuR) in spliceosomes assembled from introns containing miR-18 and miR-19a [18]. Although it is not an integral component of the spliceosome, HuR is associated with splicing proteins and the RNA interference machinery, such as the Argonaute proteins [19]. It is also engaged in miRNA processing steps [20].
HuR, also known as embryonic lethal abnormal vision-like 1 (ELAVL1), is a ubiquitously expressed RNA-binding protein (RBP) composed of three RNA recognition motifs (RRMs) [21, 22]. Approximately 90% of endogenous HuR is concentrated in the nucleus, where it has a role in RNA processing, especially in polyadenylation and splicing [21-24]. HuR is overexpressed in many cancer types and has been implicated in regulating the cell cycle, tumorigenesis (cell proliferation, migration, and invasion), immunity, and angiogenesis [24-28]. Silencing HuR using RNAi reduced proliferation, migration, and invasion of ovarian tumor cells [29]. In addition, HuR has been related to the regulation of vascular endothelial growth factor A (VEGFA) expression and angiogenesis [30], also being associated with the inflammatory process in human macrophages [31]. Previous studies reported that HuR could either control the target sites on the 3’UTR of its own mRNA or compete with miRNAs to bind the 3’UTR of other targets [30, 32, 33].
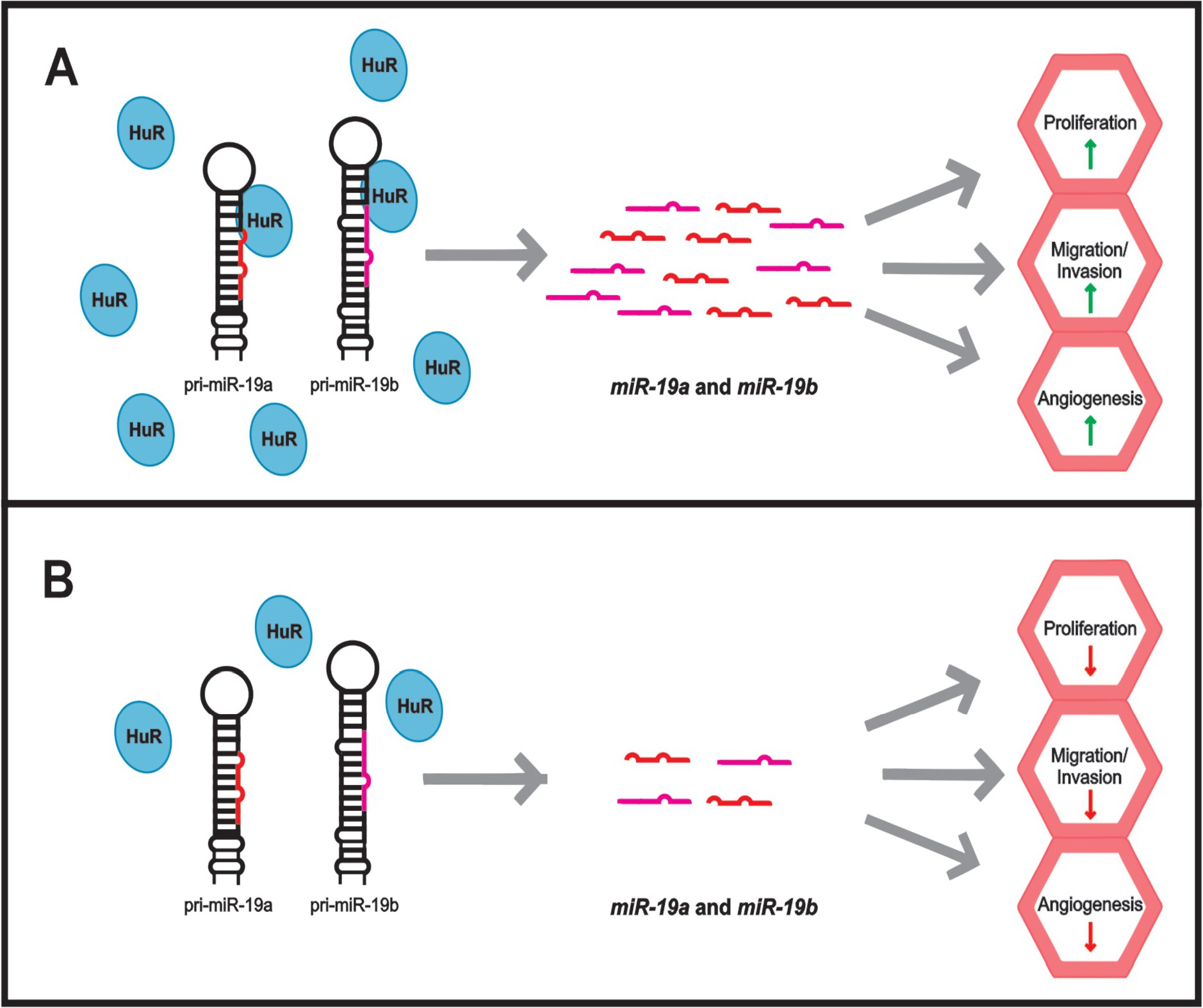
We hypothesized that HuR could associate with miRNAs of this cluster and participate on the processing steps of this intron, therefore controlling miRNA biogenesis and maturation. As a consequence, it would be a key regulator of tumorigenesis processes triggered by these miRNAs. To address that, we investigated the biogenesis and maturation of these miRNAs in papillary thyroid cancer cell line (BCPAP) under altered expression of HuR. Importantly, we observed HuR associates and affects the expression of miR-19a and miR-19b . We also confirmed phenotypic alterations in cells over-expressing this protein, indicating a possible role in thyroid cancer development.
Materials and Methods
Cell culture
HeLa-Cre, HEK-293T, and papillary thyroid cancer (BCPAP) cell lines were maintained in DMEM/high-glucose (Thermo Fisher Scientific) supplemented with 10% FBS (HyClone), 1 mM sodium pyruvate (Life Technologies), L-glutamine, and 1X penicillin-streptomycin (100 U/mL penicillin, 100 μg/mL streptomycin; Life Technologies) in 60 mm Petri dishes, unless otherwise indicated. Adherent cells were detached using 1X trypsin/EDTA (Life Technologies). Cells were cultured at 37 °C in a humidified, controlled atmosphere incubator (95% air, 5% CO2). According to the manufacturer’s instructions, transfections were performed with Lipofectamine 2000 (Life Technologies). HeLa-Cre was kindly provided by E. Makeyev [34], and BCPAP was kindly provided by Massimo Santoro (University “Federico II”, Naples, Italy). Transfection selection was performed by gradually increasing geneticin (G418, Sigma) concentration to 1000 µg/mL, generating stably transfected cells. Cells were maintained in geneticin at 200 µg/mL.
microRNA quantification (qRT-PCR) and TaqMan-Based PCR
Whole-cell extracts were prepared using buffer A (10 mM KCl, 1.5 mM MgCl2, 20 mM HEPES [pH 7.5], 0.5 mM DTT) and the Douncer homogenizer (Wheaton, NJ). RNA was extracted using Trizol reagent (Thermo Fisher Scientific) and precipitated with sodium acetate and ethanol. According to the manufacturer’s instructions, this material was used for cDNA synthesis using Superscript IV RT enzyme (Life Technologies) and random primers. 100 ng of these cDNAs were used in real-time RT-PCR reactions (qRT-PCR) using SYBR Green reagent (Thermo Fisher Scientific) and specific primers for miRNAs 17a, 18a, 19a and 92a, RNU6B, HuR, and b-actin (Supplementary Table S1 – for all supplementary material see www.cellphysiolbiochem.com). TaqMan analyses were performed using 200 nM of probes for hsa-miR-19a, hsa-miR-19b, hsa-miR-18, and hsa-miR-423 (Thermo Fisher Scientific) (Supplementary Table S1). The probes for hsa-miR-423 were used as endogenous control, as recommended by Thermo. TaqMan universal master mix (ThermoFisher Scientific) was used according to manufacturer’s instructions, and ~100 ng DNA, in a total volume of 15 μl were run with the following cycling conditions: 16 °C for 30 min, 42 °C for 30 min, 85 °C for 5 min. Expression of specific miRNAs were normalized with U6 small nuclear RNA (RNU6B) when using SYBR Green or hsa-miR-423 when using the TaqMan probe system. The fold change calculation was performed using the delta-delta Ct (2-ΔΔCt) method [35].
Immunoprecipitation
pFLAG-HuR was transfected into HEK-293T and BCPAP cells. As controls, empty pFLAG and untransfected cells were used. Cells were collected, and extracts were prepared as described above. These extracts were immunoprecipitated by incubation with protein A-Sepharose coupled to anti-FLAG M2 (Sigma) for 16 to 18 h at 4 °C. After incubation, the resin was extensively washed in RIPA buffer (20 mM Tris [pH 8.0], 150 mM of NaCl; 10% glycerol, 2 mM EDTA; 2 mM EGTA, 1% Triton X-100) and the proteins were eluted using 5 µg/µl FLAG peptide for 2 h under rotation. Input and elution fractions were subjected to RNA extraction and qRT-PCR analysis, as described.
Luciferase reporter assay
To develop the luciferase reporter assay, three plasmids were created. We inserted pri-miR-17-92 sequence into the XhoI/EcoRI sites of the pRD-RIPE intron (kindly provided by Dr. E. Makeyev, Nanyang Technological University, Singapore; [34]), creating pRD-RIPE-1792. This plasmid has a tetracycline-controlled promoter, which is reversibly turned on or off by the presence of the antibiotic doxycycline. The luciferase reporter plasmid and the “scramble” control were generated using pmir-GLO (Promega). miRbase and TargetScan predicted RAP-IB 3’UTR as a target of miR-19a and miR-19b. Full length (380 bp) and scrambled (318 bp) sequences of human RAP-IB mRNA 3’-UTR were subcloned downstream into pmir-GLO at XhoI/XbaI sites, generating pmirGLO-RAP-IB-3ʹ-UTR (Luc-RAP-IB-3’-UTR) and pmirGLO-scrambled-3ʹ-UTR (Luc-scrambled-3’-UTR) reporter constructs. The sequence and orientation of the luciferase reporter were verified by NotI cleavage and DNA sequencing. To perform the reporter assay, HeLa-Cre cells were co-transfected with pRD-RIPE-1792 (300 ng) and pCAGGS-Cre (Cre-encoding plasmid) (100 ng) (kindly provided by Dr. E. Makeyev, Nanyang Technological University, Singapore; [34]). Co-transfection was performed using confluent cells on a 24-well plate. These cells were incubated with a mixture containing 2.0 μg of DNA and 1.25 μL Lipofectamine 2000 in 100 μL Opti-MEM I (Life Technologies, Carlsbad, CA) following the manufacturer’s protocol. Cells were incubated with the transfection mixtures overnight, the medium was replaced for DMEM, and the incubation continued for another 24 h before adding puromycin. These cells were then used for the transfection of the luciferase reporter plasmid. This was performed with 50 ng/mL of each Luc-RAP-IB-3’-UTR and Luc-scrambled-3’-UTR, separately, using Lipofectamine 2000 as recommended (Life Technologies, Carlsbad, CA). The medium was replaced 6-8 h post-transfection to include the mixture of penicillin and streptomycin, and the incubation continued for another 20 h in DMEM. The two luciferases’ activity was measured 48 h post-transfection using the Dual-Glo Luciferase Assay System (Promega).
Small Interfering RNA (siRNA) and antimiR-19a transfection
Silencer selected validated small interfering RNA (siRNA) for HuR (catalogue number 4390824) and negative control siRNA (catalogue number 4390843) were purchased from Life Technologies (Supplementary Table S1). To inhibit HuR expression, three different siRNA concentrations were transfected into BCPAP cells using Lipofectamine 2000 (2.5, 5, and 10 nM) (Life Technologies), following the manufacturer’s protocol. Cells were collected after 24 h and subjected to protein preparation and RNA extraction. 25 pM of antimiR-19a (miRVana, Thermo catalogue number 4464084) and the negative control (mirVana, Thermo catalogue number 4464076) (Supplementary Table S1) were transfected into BCPAP-FLAG and BCPAP-HuR cells using Lipofectamine 2000 (Life Technologies) according to manufacturer’s protocol. Cell growth after 24h and 72h post-transfection was evaluated using trypan blue.
Western Blot
Proteins were extracted using a buffer composed of 10 mM Tris-HCl pH 8, 140 mM NaCl, 0.1% SDS, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM EDTA, 0.5 mM EGTA, 1 mM PMSF and 1 mM DTT. Total protein concentrations were determined using the Bradford reagent (BioRad). Equal amounts of samples prepared from whole-cell extracts were separated on 10% SDS-PAGE, transferred to a nitrocellulose membrane, and probed with anti-HuR rabbit monoclonal (Cell Signaling) and anti-b-actin (Sigma). Following incubation with secondary antibodies IRDye 680/800CW-labeled rabbit or mouse (LI-COR Bioscience), blots were visualized using Odyssey CLx imaging system software (LI-COR Bioscience). Quantification was performed using Image J software and depicted after normalization of optic densitometry.
Cell growth, migration, and invasion
Cell growth was evaluated for 48 h with trypan blue staining. BCPAP cells at an initial concentration of 2.5 x 104/mL were cultured in 6-well plates (Corning) at 37 °C in 5% CO2 for 48 h. After cell suspension, a 0.4% trypan blue solution (Sigma Aldrich) was added in a 1:1 ratio. After 3 min, cells were counted and separated into live cells (no cytoplasmic fluorescence) and dead cells (blue cytoplasmic fluorescence). The trypan blue-positive ratio from 10 random fields was quantified with ImageJ software. Cells were counted using the Countess II FL (Life Technologies). Migration and invasion assays were performed using 8.0 μm pore transwell membranes (Corning). Membranes were incubated with PBS for 1 h at 37 °C, 5 % CO2 atmosphere for migration assays. For invasion assays, membranes were coated with 25 μg Matrigel® (BD Biosciences) and incubated for 1 h at 37 °C, 5% CO2 atmosphere. For both assays, about 2.5 x 104/mL cells were suspended in a culture medium containing 1 % FBS and plated in the upper chamber, whereas the lower chamber had 10 % FBS supplemented media. Non-migrating cells were removed from the top chamber after 24h using a cotton swab. Migrating cells were fixed with 4 % paraformaldehyde (PFA) in PBS and stained with 0.5 % Crystal Violet. Cells were photographed using a Nikon Eclipse E600 microscope equipped with optical camera CF160 epifluorescence, and ten representative fields were counted. Total protein concentrations were determined using the Bradford reagent.
Statistical analyses
Data are presented as mean ± SEM. Statistical analyses were performed using GraphPad Prism (GraphPad software, version 9, San Diego). Splicing reporter assay results and immunoprecipitation results were analyzed by two-way ANOVA followed by Tukey’s post-test to allow group comparison. Results of qPCR and Taqman experiments were further analyzed using the Mann-Whitney post-test. Differences at p-values <0.05 were considered to be significant.
Results
HuR overexpression in BCPAP and HEK-293T cells
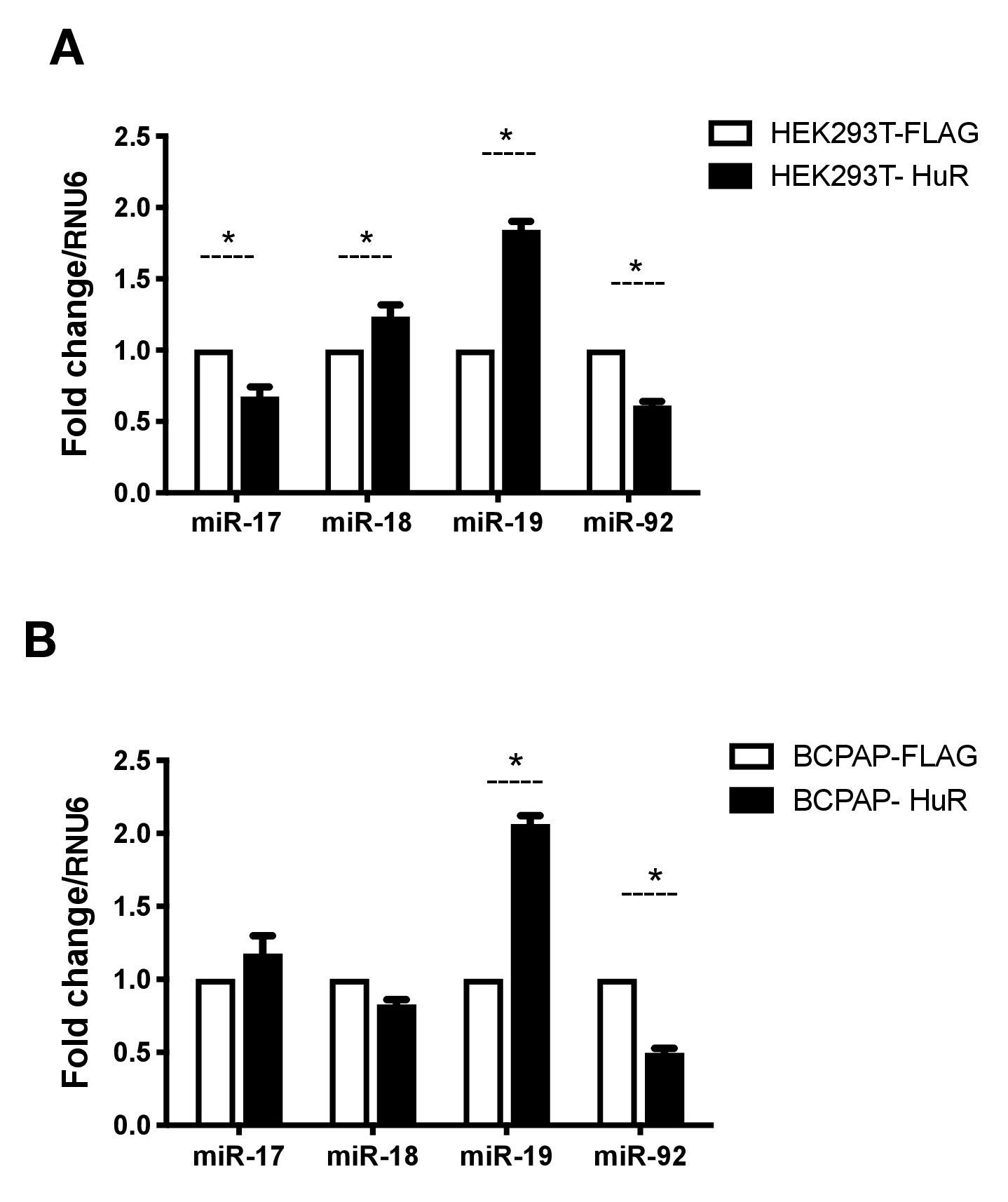
We previously observed HuR in spliceosomes assembled upon introns containing miR-18a and miR-19a [18]. To investigate whether HuR could modulate the expression of miR-17-92 miRNAs in cancer cells, we induced HuR overexpression in vitro in two cell lines: human embryonic kidney (HEK-293T) and papillary thyroid cancer (BCPAP). BCPAP is a papillary thyroid cell line carrying the mutation BRAFV600E [7]. HEK-293T is derived from human embryonic kidney. Previous genomic analysis revealed HEK-293T has 7 copies of MIR17HG [36]. We reasoned that HuR regulatory roles on the expression of the miRNAs could be compared in these two cell lines. HuR overexpression in BCPAP and HEK-293T cells was confirmed by real-time PCR (Supplementary Fig. 1). We first analyzed miR-17-92 individual levels after HuR overexpression. As a control, empty pFLAG was also transfected into these cells. BCPAP-HuR and HEK293T-HuR showed increased expression of miR-19a , suggesting this protein positively regulates miR-19a expression independently of the cell line. On the other hand, we observed reduced levels of miR-92a in both cell lines (Fig. 1). The evidence for the role of HuR on the regulation of these miRNAs is reinforced by the fact that multiple copies of this cluster are found in HEK-293T cells.
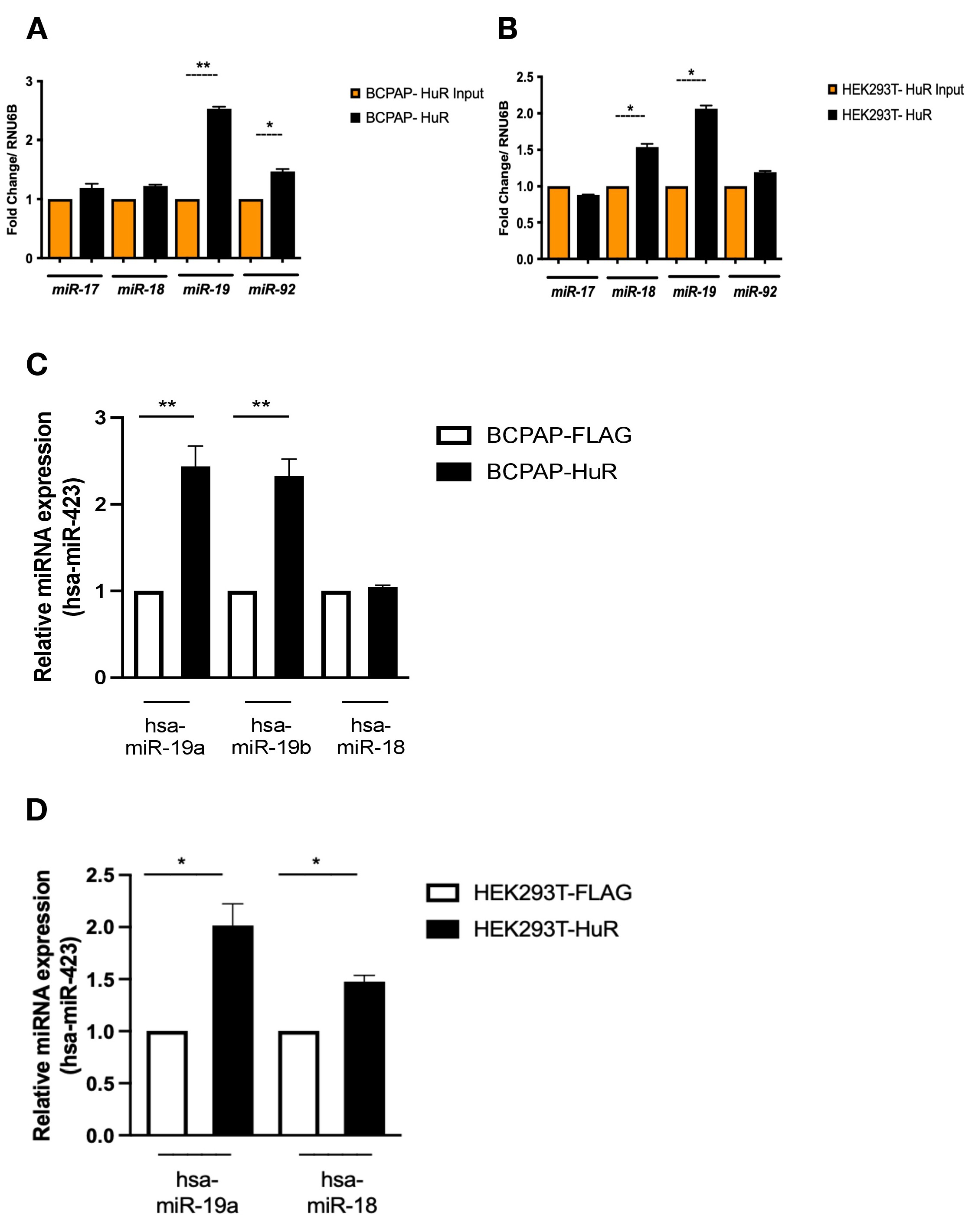
Our qPCR using HuR over-expressing cells and in silico analysis retrieved from miRbase and TargetScan databases using query search consistently revealed potential HuR binding sites along pre-miR-17-92 sequence (GUUU, AUGA, NNUUNNUUU). Specifically, pre-miR-19a and pre-miR-19b sequences show conserved HuR binding sites (Fig. 2), indicating a possible interaction with these regions of the cluster. To investigate the association of HuR with this region, we performed immunoprecipitation using HuR over-expressing cells. Whole-cell extracts from BCPAP-HuR, HEK293T-HuR, and controls (cells expressing only the epitope FLAG) were subjected to anti-FLAG immunoprecipitation, and input and elution fractions were analyzed using qPCR. With three biological replicates, we observed that levels of miR-19a and miR-92a increased in BCPAP-HuR elution fractions relative to controls, indicating an association of those miRNAs and HuR (Fig. 3A). HuR associates with miR-19a in HEK293T-HuR as well (Fig. 3B). Specifically, HuR is also associated with miR-18 in this cell line. With the use of Taqman probes, we also confirmed strong HuR association with hsa-miR-19a and hsa-miR19b in BCPAP (Fig. 3C) and in HEK-293T cells (Fig. 3D). Specific Taqman probes for hsa-miR18 also confirmed association with HuR in HEK-293T but not in BCPAP cells (Fig. 3C and 3D).

Knockdown of HuR affects the synthesis of miR-19a and miR-17-5p
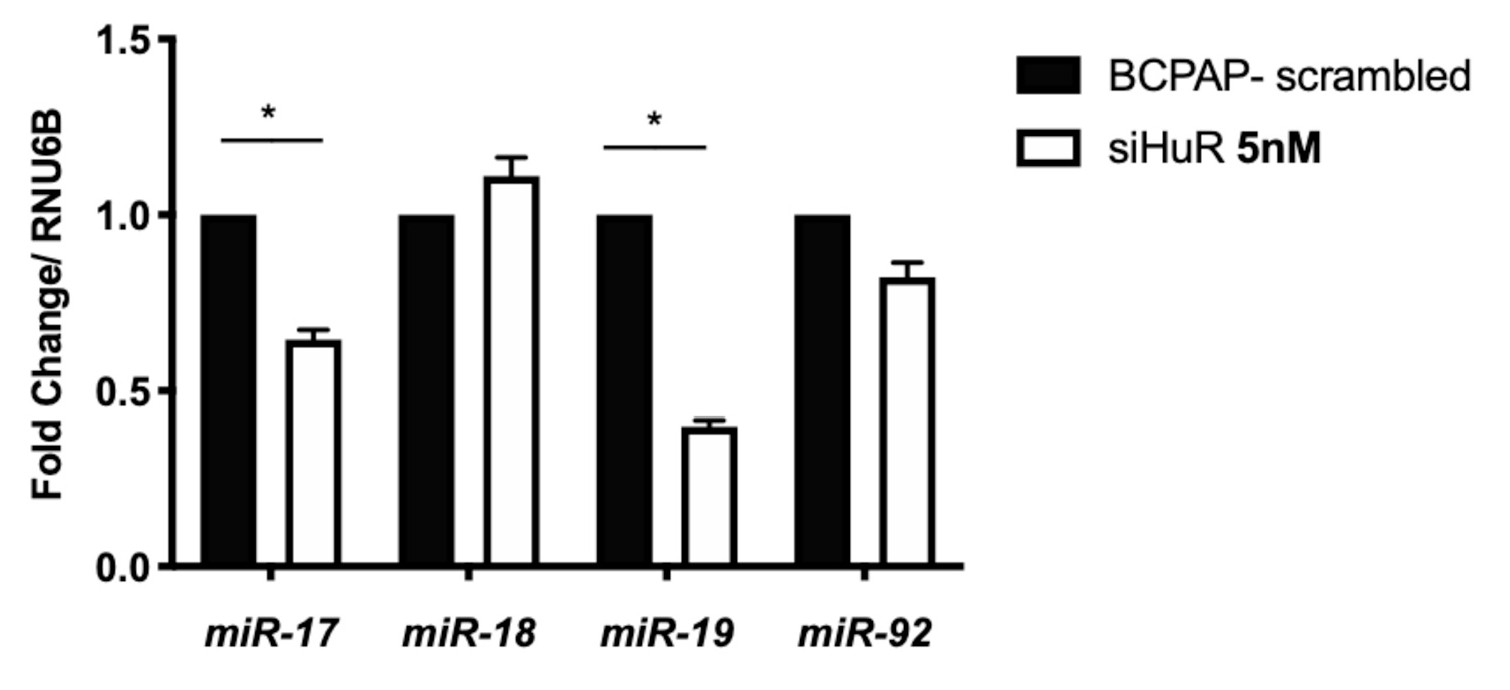
Considering HuR was associated with miR-19a and miR-19b , we asked whether its absence would also interfere with the expression of these miRNAs in BCPAP cells. To reduce the levels of HuR, we transfected BCPAP cells with siRNA against HuR (siHuR). Concentrations as low as 5 nM led to reduced HuR mRNA and protein, as confirmed by qPCR and western blot against HuR (Supplementary Fig. 2). We then analyzed the expression of miRNAs in BCPAP-siHuR cells by qRT-PCR. Our results indicated that miR-17-5p and miR-19a levels decreased significantly in BCPAP-siHuR cells (Fig. 4). miR-18 and miR-92 remained unchanged after using siHuR in BCPAP cells. Therefore, the reduction observed in miR-19a might be primarily due to the strong association of HuR with miR-19a , as observed with the IP assay (Fig. 3). Additionally, we also observe a reduction in miR-17-5p upon knockdown of HuR. Since HuR can bind next to miR-17-5p coding region, it is possible that its absence also impacts the processing of this miRNA. This result indicates that HuR not only associates with miR-19a but also regulates the expression miR-19a and miR-17-5p. Thus, it is possible that HuR binding to the pre-miRNA facilitates the processing of the miRNAs transcribed from the 5’-end of the cluster, such as miR-17-5p, miR-18 and miR-19a [37].
HuR induces miR-19 expression and maturation
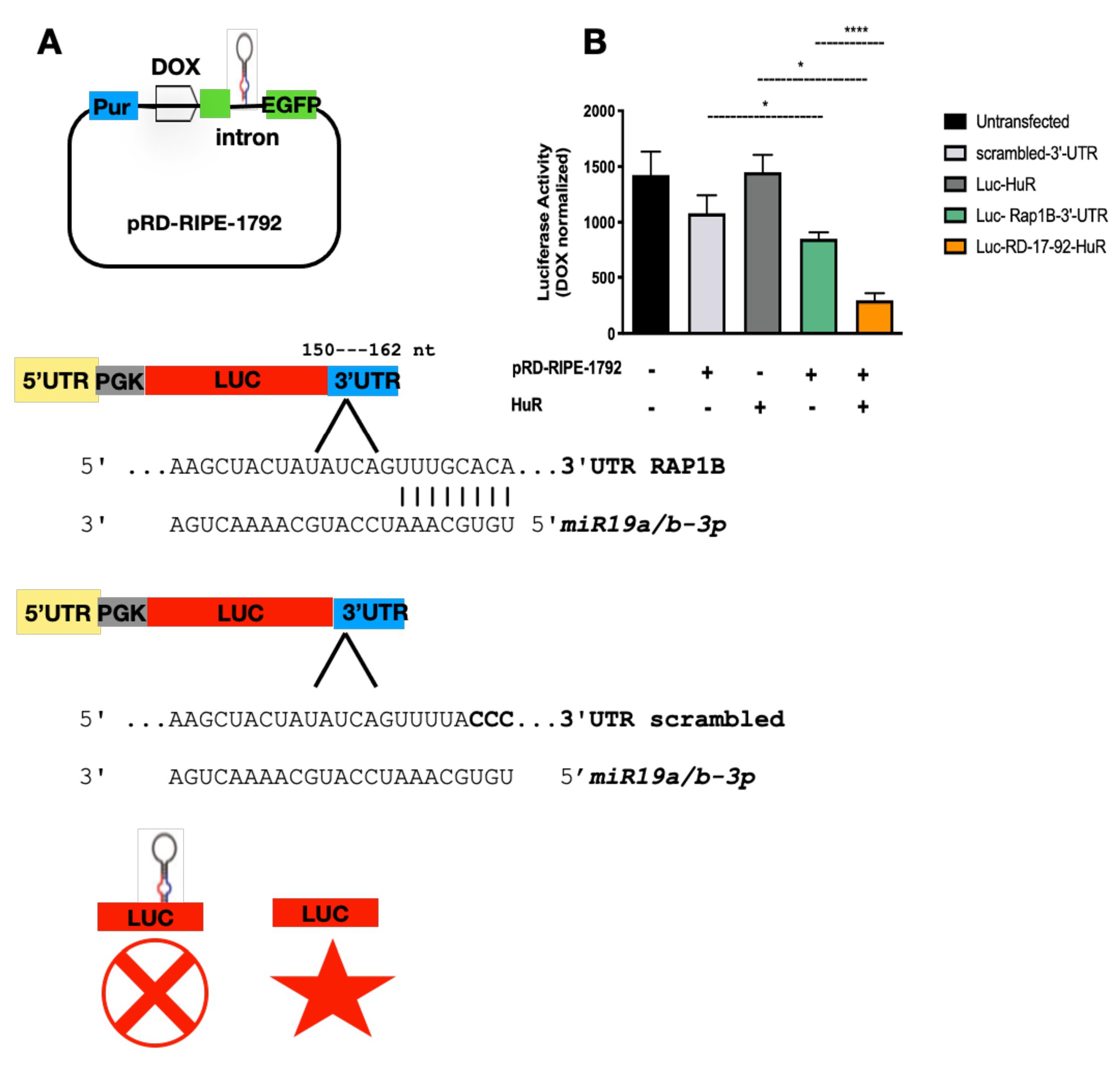
Our results indicated that HuR binds to and regulates the expression of miR-19a and miR-19b in BCPAP cells. The connection of HuR expression and miR-17-92 cluster could point to a new mechanism governing the biogenesis of these miRNAs. To confirm that the induced miRNAs were truly functional, we designed an in vitro reporter system using a plasmid containing intronic pri-miR-17-92, whose expression is controlled by a tetracycline-inducible promoter (Fig. 5A) [34]. Ras-related protein RAP-IB is a GTPase member of the Ras-associated protein family (RAS). Bioinformatics analysis revealed that RAP-IB 3’ UTR has target sequences for both miR-19a and miR-19b. Therefore, this sequence was inserted into pmiR-GLO to test for miR-19a and miR-19b activity. If the induced miRNA were correctly processed and functional in our assay, it would hybridize to the sequence cloned next to luciferase. Therefore, it would block luciferase translation, resulting in reduced luciferase activity. Under the absence of doxycycline, and therefore without activation of exogenous miR-17-92 expression, we observed endogenous miR-19a and/or miR-19b successfully found the target, resulting in reduced luciferase expression and activity (Luc-RAP-IB-3’UTR). The addition of FLAG-HuR coupled to doxycycline supplementation further reduced luciferase activity, indicating that more miR-19a and miR-19b were synthesized and able to find their target in pmiR-GLO-RAP-IB (Fig. 5B, Luc-RD-17-92-HuR). Altogether, these results support that induction by HuR generated functional miR-19a and miR-19b.
HuR overexpression is associated with tumor progression
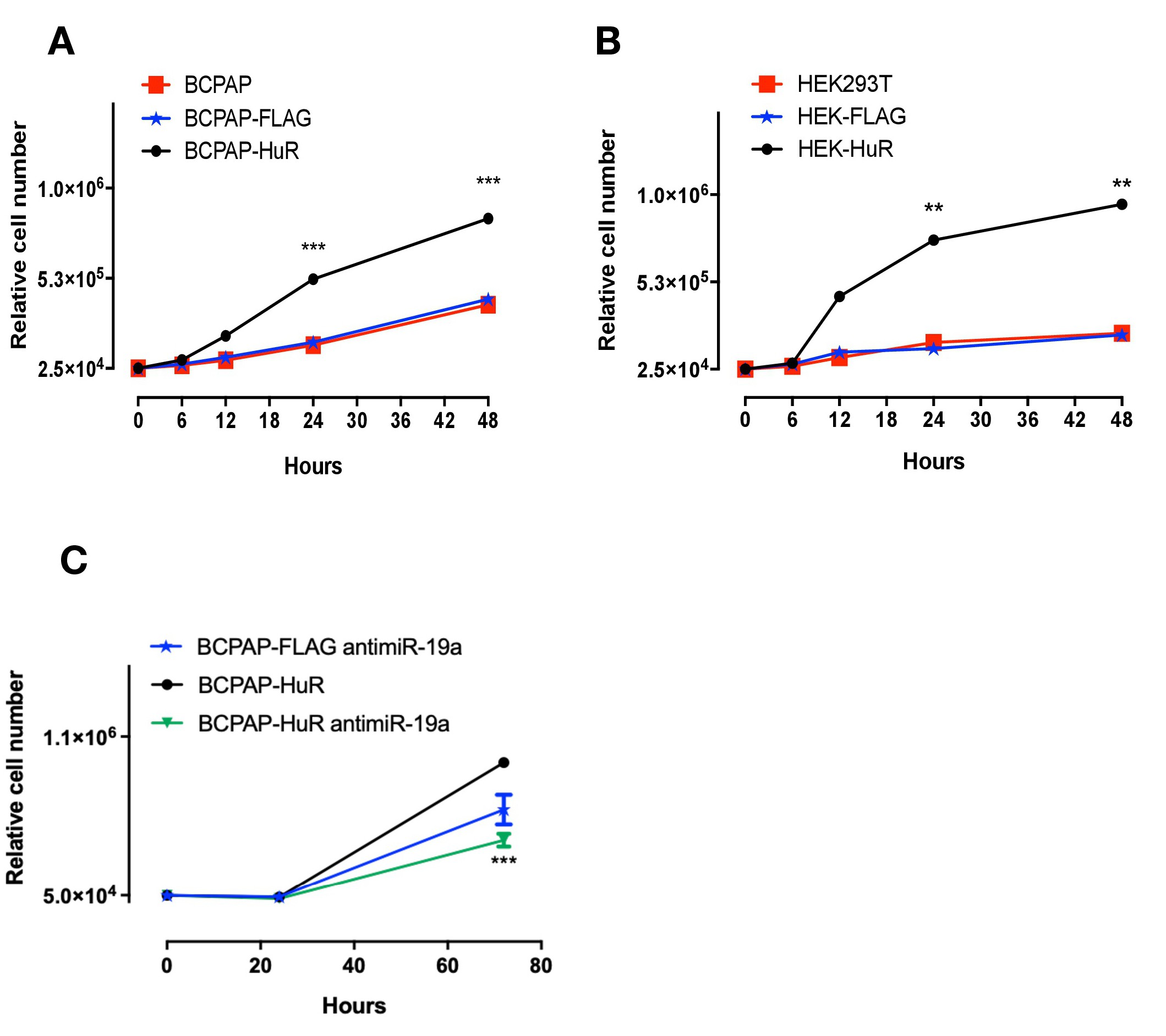
Our results indicated that HuR positively controls miR-19a and miR-19b biogenesis and processing, and these miRNAs were already shown as potent oncogenic miRNAs [8]. We then hypothesized that increased expression of HuR could affect cellular kinetics promoting tumorigenic effects. To evaluate that, we analyzed cell growth, migration and invasion in cells over-expressing HuR. By comparing the phenotypes of BCPAP-HuR and BCPAP-FLAG; and HEK293T-HuR and HEK293T-FLAG, we observed increased growth in BCPAP-HuR and HEK293T-HuR cells (Fig. 6A and 6B), suggesting HuR is stimulating cell growth. We reasoned that HuR was stimulating miR-19a expression, which was the responsible for the increased cell growth observed. To address that, we transfected BCPAP-FLAG and BCPAP-HuR cells with antimiR-19a or with a negative control antimiRNA. Following 24h of the transfection, we confirmed miR-19a inhibition by qRT-PCR (Supplementary Fig. 3). We then analyzed cell growth for another 48h and compared it with the growth of BCPAP-HuR cells. The results showed inhibition of miR-19a has a significant impact on growth of these cells, especially when compared to BCPAP-HuR cells (P<0.0005) (Fig. 6C).
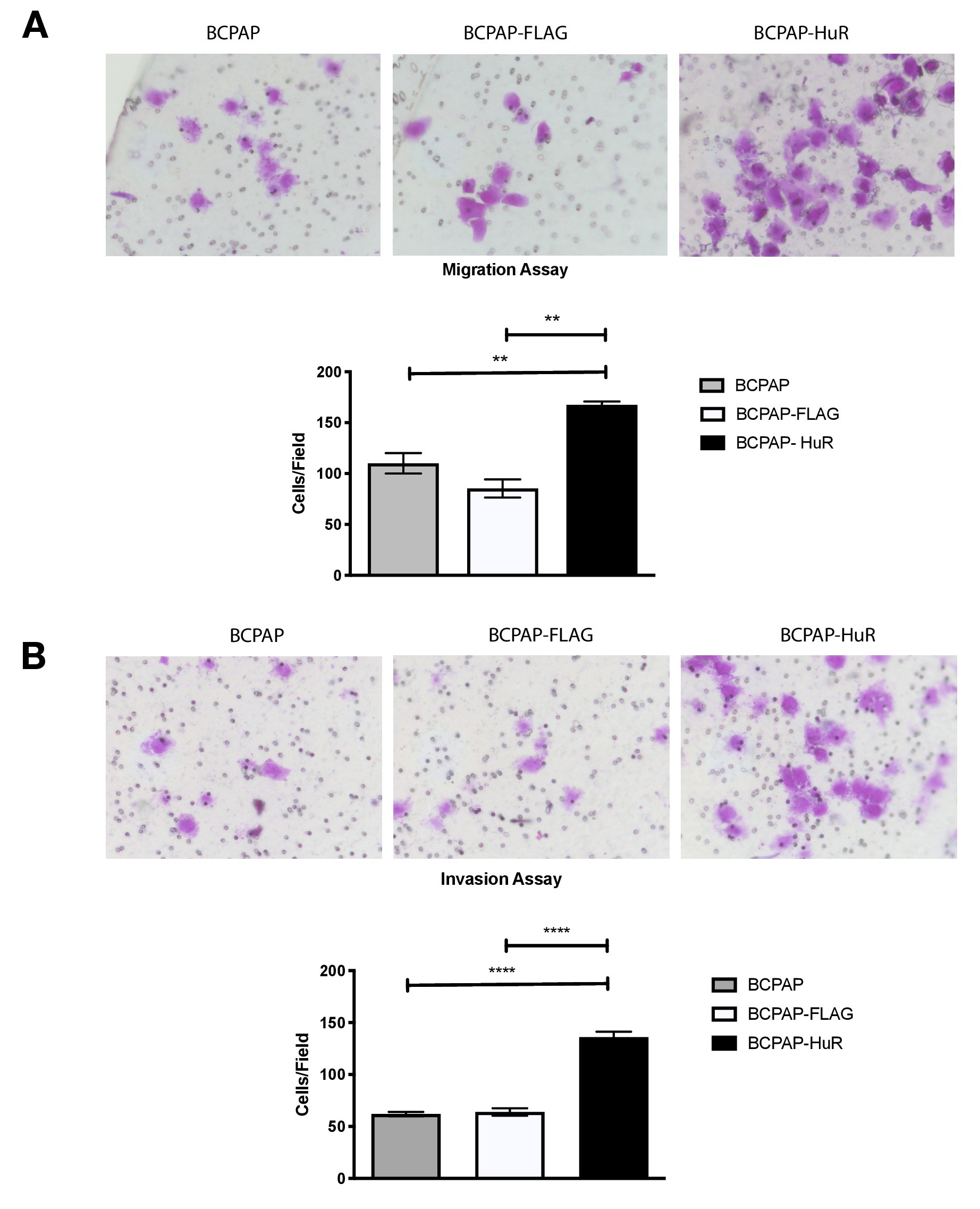
Sustained continuous proliferation is one hallmark of cancer [38], and this characteristic indicates that HuR over-expression could affect other tumorigenic aspects. We then sought to investigate migration and invasion rates in cells overexpressing HuR. Transwell migration assays were performed with approximately 2.5 x 104 cells/mL after incubation of 24 h in regular culture media containing FBS. Our results showed that BCPAP cells overexpressing HuR migrated faster than control cells. In addition, the number of cells counted on the lower chamber was, on average, 50% higher than the number found in control cells (Fig. 7A).
Similarly, we performed this assay using a matrigel layer over the transwell membrane, simulating the extracellular matrix environment. The number of HuR overexpressing cells that could invade the lower chamber was twice the number found for control cells (Fig. 7B). This result indicated that HuR over-expressing cells also had increased invasive capacity. Altogether, these results suggested that HuR over-expression increases tumorigenic characteristics in thyroid papillary cancer cell line. Indeed, miR-19a and miR-19b stimulation by HuR over-expression might directly involve these observed effects during tumor progression.
We are grateful to Eugene Makeyev, Edna T. Kimura and Melissa Jurica for kindly providing reagents. We are also thankful to Cilene Rebouças, Marinilce Fagundes dos Santos, Lucia Rosetti Lopes, Glaucia Santelli, César Fuziwara, Vanessa Freitas, Carolina Purcell, and Gisela Ramos for their support.
Author Contributions
Conception of the work: G.H.G.S. and P.P.C.; Acquisition, analysis and interpretation of data: G.H.G.S., M.G.P.S., H.Y.N. and P.P.C.; Manuscript draft: G.H.G.S. and P.P.C.; Manuscript final revision: G.H.G.S., H. Y. N., M.G.P.S. and P.P.C.; Funding: P.P.C.
Funding
G.H.G.S., M.G.P.S. and H.Y.N. received fellowships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior; P.P.C. was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo, Grant Numbers 2017/06994-9 and 2019/21874-5.
Statement of Ethics
The authors have no ethical conflicts to disclose.
The authors declare that no conflicts of interest exist.
| 1 Matera AG, Wang Z: A day in the life of the spliceosome. Nat Rev Mol Cell Biol 2014;15:108-121. https://doi.org/10.1038/nrm3742 |
||||
| 2 Coltri PP, Dos Santos MGP, da Silva GHG: Splicing and cancer: Challenges and opportunities. Wiley Interdiscip Rev RNA RNA 2019;10:e1527. https://doi.org/10.1002/wrna.1527 |
||||
| 3 Franca GS, Vibranovski MD, Galante PA: Host gene constraints and genomic context impact the expression and evolution of human microRNAs. Nat Commun 2016;7:11438. https://doi.org/10.1038/ncomms11438 |
||||
| 4 Lin SL, Ying SY: Mechanism and Method for Generating Tumor-Free iPS Cells Using Intronic MicroRNA miR-302 Induction. Methods Mol Biol 2018;1733:265-282. https://doi.org/10.1007/978-1-4939-7601-0_22 |
||||
| 5 Bartel DP: MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-233. https://doi.org/10.1016/j.cell.2009.01.002 |
||||
| 6 Friedman RC, Farh KK, Burge CB, Bartel DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105. https://doi.org/10.1101/gr.082701.108 |
||||
| 7 Fuziwara CS, Kimura ET: High iodine blocks a Notch/miR-19 loop activated by the BRAF(V600E) oncoprotein and restores the response to TGFbeta in thyroid follicular cells. Thyroid 2014;24:453-462. https://doi.org/10.1089/thy.2013.0398 |
||||
| 8 Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L: miR-19 is a key oncogenic component of mir-17-92. Genes Dev 2009;23:2839-2849. https://doi.org/10.1101/gad.1861409 |
||||
| 9 Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T: A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 2005;65:9628-9632. https://doi.org/10.1158/0008-5472.CAN-05-2352 |
||||
| 10 Bai X, Hua S, Zhang J, Xu S: The MicroRNA Family Both in Normal Development and in Different Diseases: The miR-17-92 Cluster. Biomed Res Int 2019;2019:9450240. https://doi.org/10.1155/2019/9450240 |
||||
| 11 Olive V, Jiang I, He L: mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol 2010;42:1348-1354. https://doi.org/10.1016/j.biocel.2010.03.004 |
||||
| 12 Abasi M, Kohram F, Fallah P, Arashkia A, Soleimani M, Zarghami N, Ghanbarian H: Differential Maturation of miR-17 ~ 92 Cluster Members in Human Cancer Cell Lines. Appl Biochem Biotechnol 2017;182:1540-1547. https://doi.org/10.1007/s12010-017-2416-5 |
||||
| 13 Dews M, Fox JL, Hultine S, Sundaram P, Wang W, Liu YY, Furth E, Enders GH, El-Deiry W, Schelter JM, Cleary MA, Thomas-Tikhonenko A: The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res 2010;70:8233-8246. https://doi.org/10.1158/0008-5472.CAN-10-2412 |
||||
| 14 Mestdagh P, Bostrom AK, Impens F, Fredlund E, Van Peer G, De Antonellis P, von Stedingk K, Ghesquiere B, Schulte S, Dews M, Thomas-Tikhonenko A, Schulte JH, Zollo M, Schramm A, Gevaert K, Axelson H, Speleman F, Vandesompele J: The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol Cell 2010;40:762-773. https://doi.org/10.1016/j.molcel.2010.11.038 |
||||
| 15 Takakura S, Mitsutake N, Nakashima M, Namba H, Saenko VA, Rogounovitch TI, Nakazawa Y, Hayashi T, Ohtsuru A, Yamashita S: Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci 2008;99:1147-1154. https://doi.org/10.1111/j.1349-7006.2008.00800.x |
||||
| 16 Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M: HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 2009;23:1743-1748. https://doi.org/10.1101/gad.1812509 |
||||
| 17 Thomas M, Lange-Grunweller K, Hartmann D, Golde L, Schlereth J, Streng D, Aigner A, Grunweller A, Hartmann RK: Analysis of transcriptional regulation of the human miR-17-92 cluster; evidence for involvement of Pim-1. Int J Mol Sci 2013;14:12273-12296. https://doi.org/10.3390/ijms140612273 |
||||
| 18 Paiva MM, Kimura ET, Coltri PP: miR18a and miR19a Recruit Specific Proteins for Splicing in Thyroid Cancer Cells. Cancer Genomics Proteomics 2017;14:373-381. https://doi.org/10.21873/cgp.20047 |
||||
| 19 Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G: Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 2007;8:1052-1060. https://doi.org/10.1038/sj.embor.7401088 |
||||
| 20 Manzoni L, Zucal C, Maio DD, D'Agostino VG, Thongon N, Bonomo I, Lal P, Miceli M, Baj V, Brambilla M, Cerofolini L, Elezgarai S, Biasini E, Luchinat C, Novellino E, Fragai M, Marinelli L, Provenzani A, Seneci P: Interfering with HuR-RNA Interaction: Design, Synthesis and Biological Characterization of Tanshinone Mimics as Novel, Effective HuR Inhibitors. J Med Chem 2018;61:1483-1498. https://doi.org/10.1021/acs.jmedchem.7b01176 |
||||
| 21 Chae MJ, Sung HY, Kim EH, Lee M, Kwak H, Chae CH, Kim S, Park WY: Chemical inhibitors destabilize HuR binding to the AU-rich element of TNF-alpha mRNA. Exp Mol Med 2009;41:824-831. https://doi.org/10.3858/emm.2009.41.11.088 |
||||
| 22 Lu YC, Chang SH, Hafner M, Li X, Tuschl T, Elemento O, Hla T: ELAVL1 modulates transcriptome-wide miRNA binding in murine macrophages. Cell Rep 2014;9:2330-2343. https://doi.org/10.1016/j.celrep.2014.11.030 |
||||
| 23 Dai W, Zhang G, Makeyev EV: RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Res 2012;40:787-800. https://doi.org/10.1093/nar/gkr783 |
||||
| 24 Lee JH, Jung M, Hong J, Kim MK, Chung IK: Loss of RNA-binding protein HuR facilitates cellular senescence through posttranscriptional regulation of TIN2 mRNA. Nucleic Acids Res 2018;46:4271-4285. https://doi.org/10.1093/nar/gky223 |
||||
| 25 Katsanou V, Milatos S, Yiakouvaki A, Sgantzis N, Kotsoni A, Alexiou M, Harokopos V, Aidinis V, Hemberger M, Kontoyiannis DL: The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol Cell Biol 2009;29:2762-2776. https://doi.org/10.1128/MCB.01393-08 |
||||
| 26 Abdelmohsen K, Gorospe M: Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA 2010;1:214-229. https://doi.org/10.1002/wrna.4 |
||||
| 27 Turner M, Diaz-Munoz MD: RNA-binding proteins control gene expression and cell fate in the immune system. Nat Immunol 2018;19:120-129. https://doi.org/10.1038/s41590-017-0028-4 |
||||
| 28 Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y, Li H, Chen Y, Wang X, Huang K, Zheng L, Tong Q: Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer 2019;18:158. https://doi.org/10.1186/s12943-019-1094-z |
||||
| 29 Huang YH, Peng W, Furuuchi N, Gerhart J, Rhodes K, Mukherjee N, Jimbo M, Gonye GE, Brody JR, Getts RC, Sawicki JA: Delivery of Therapeutics Targeting the mRNA-Binding Protein HuR Using 3DNA Nanocarriers Suppresses Ovarian Tumor Growth. Cancer Res 2016;76:1549-1559. https://doi.org/10.1158/0008-5472.CAN-15-2073 |
||||
| 30 Chang SH, Lu YC, Li X, Hsieh WY, Xiong Y, Ghosh M, Evans T, Elemento O, Hla T: Antagonistic function of the RNA-binding protein HuR and miR-200b in post-transcriptional regulation of vascular endothelial growth factor-A expression and angiogenesis. J Biol Chem 2013;288:4908-4921. https://doi.org/10.1074/jbc.M112.423871 |
||||
| 31 Zhang J, Modi Y, Yarovinsky T, Yu J, Collinge M, Kyriakides T, Zhu Y, Sessa WC, Pardi R, Bender JR: Macrophage beta2 integrin-mediated, HuR-dependent stabilization of angiogenic factor-encoding mRNAs in inflammatory angiogenesis. Am J Pathol 2012;180:1751-1760. https://doi.org/10.1016/j.ajpath.2011.12.025 |
||||
| 32 Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I: A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell 2014;53:506-514. https://doi.org/10.1016/j.molcel.2013.12.012 |
||||
| 33 Chang N, Ge J, Xiu L, Zhao Z, Duan X, Tian L, Xie J, Yang L, Li L: HuR mediates motility of human bone marrow-derived mesenchymal stem cells triggered by sphingosine 1-phosphate in liver fibrosis. J Mol Med (Berl) 2017;95:69-82. https://doi.org/10.1007/s00109-016-1460-x |
||||
| 34 Khandelia P, Yap K, Makeyev EV: Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc Natl Acad Sci U S A 2011;108:12799-12804. https://doi.org/10.1073/pnas.1103532108 |
||||
| 35 Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-408. https://doi.org/10.1006/meth.2001.1262 |
||||
| 36 Lin YC, Boone M, Meuris L, Lemmens I, Van Roy N, Soete A, Reumers J, Moisse M, Plaisance S, Drmanac R, Chen J, Speleman F, Lambrechts D, Van de Peer Y, Tavernier J, Callewaert N: Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations. Nat Commun 2014;5:4767. https://doi.org/10.1038/ncomms5767 |
||||
| 37 Du P, Wang L, Sliz P, Gregory RI: A Biogenesis Step Upstream of Microprocessor Controls miR-17~92 Expression. Cell 2015;162:885-899. | ||||
| 38 Hanahan D, Weinberg RA: Hallmarks of cancer: the next generation. Cell 2011;144:646-674. https://doi.org/10.1016/j.cell.2011.02.013 |
||||
| 39 Baldan F, Mio C, Allegri L, Conzatti K, Toffoletto B, Puppin C, Radovic S, Vascotto C, Russo D, Di Loreto C, Damante G: Identification of tumorigenesis-related mRNAs associated with RNA-binding protein HuR in thyroid cancer cells. Oncotarget 2016;7:63388-63407. https://doi.org/10.18632/oncotarget.11255 |
||||
| 40 Aguado A, Rodriguez C, Martinez-Revelles S, Avendano MS, Zhenyukh O, Orriols M, Martinez-Gonzalez J, Alonso MJ, Briones AM, Dixon DA, Salaices M: HuR mediates the synergistic effects of angiotensin II and IL-1beta on vascular COX-2 expression and cell migration. Br J Pharmacol 2015;172:3028-3042. https://doi.org/10.1111/bph.13103 |
||||
| 41 Allegri L, Mio C, Russo D, Filetti S, Baldan F: Effects of HuR downregulation on anaplastic thyroid cancer cells. Oncol Lett 2018;15:575-579. https://doi.org/10.3892/ol.2017.7289 |
||||
| 42 Chaulk SG, Thede GL, Kent OA, Xu Z, Gesner EM, Veldhoen RA, Khanna SK, Goping IS, MacMillan AM, Mendell JT, Young HS, Fahlman RP, Glover JN: Role of pri-miRNA tertiary structure in miR-17~92 miRNA biogenesis. RNA Biol 2011;8:1105-1114. https://doi.org/10.4161/rna.8.6.17410 |
||||
| 43 Pabis M, Popowicz GM, Stehle R, Fernandez-Ramos D, Asami S, Warner L, Garcia-Maurino SM, Schlundt A, Martinez-Chantar ML, Diaz-Moreno I, Sattler M: HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs. Nucleic Acids Res 2019;47:1011-1029. https://doi.org/10.1093/nar/gky1138 |
||||
| 44 Brennan CM, Steitz JA: HuR and mRNA stability. Cell Mol Life Sci 2001;58:266-277. https://doi.org/10.1007/PL00000854 |
||||
| 45 Nabors LB, Gillespie GY, Harkins L, King PH: HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3' untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res 2001;61:2154-2161. | ||||
| 46 Galban S, Kuwano Y, Pullmann R, Jr., Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, Lewis SM, Holcik M, Gorospe M: RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol Cell Biol 2008;28:93-107. https://doi.org/10.1128/MCB.00973-07 |
||||
| 47 Zhang Z, Yao Z, Wang L, Ding H, Shao J, Chen A, Zhang F, Zheng S: Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy 2018;14:2083-2103. https://doi.org/10.1080/15548627.2018.1503146 |
||||
| 48 Zhang F, Cai Z, Lv H, Li W, Liang M, Wei X, Zhou F: Multiple functions of HuR in urinary tumors. J Cancer Res Clin Oncol 2019;145:11-18. https://doi.org/10.1007/s00432-018-2778-2 |
||||
| 49 Miyata Y, Watanabe S, Sagara Y, Mitsunari K, Matsuo T, Ohba K, Sakai H: High expression of HuR in cytoplasm, but not nuclei, is associated with malignant aggressiveness and prognosis in bladder cancer. PLoS One 2013;8:e59095. https://doi.org/10.1371/journal.pone.0059095 |
||||
| 50 Muralidharan R, Panneerselvam J, Chen A, Zhao YD, Munshi A, Ramesh R: HuR-targeted nanotherapy in combination with AMD3100 suppresses CXCR4 expression, cell growth, migration and invasion in lung cancer. Cancer Gene Ther 2015;22:581-590. https://doi.org/10.1038/cgt.2015.55 |
||||
| 51 Liang PI, Li WM, Wang YH, Wu TF, Wu WR, Liao AC, Shen KH, Wei YC, Hsing CH, Shiue YL, Huang HY, Hsu HP, Chen LT, Lin CY, Tai C, Lin CM, Li CF: HuR cytoplasmic expression is associated with increased cyclin A expression and poor outcome with upper urinary tract urothelial carcinoma. BMC Cancer 2012;12:611. https://doi.org/10.1186/1471-2407-12-611 |
||||
| 52 Carmeliet P: VEGF as a key mediator of angiogenesis in cancer. Oncology 2005;69:4-10. https://doi.org/10.1159/000088478 |
||||
| 53 Stohr N, Kohn M, Lederer M, Glass M, Reinke C, Singer RH, Huttelmaier S: IGF2BP1 promotes cell migration by regulating MK5 and PTEN signaling. Genes Dev 2012;26:176-189. https://doi.org/10.1101/gad.177642.111 |
||||
| 54 Mitsunari K, Miyata Y, Asai A, Matsuo T, Shida Y, Hakariya T, Sakai H: Human antigen R is positively associated with malignant aggressiveness via upregulation of cell proliferation, migration, and vascular endothelial growth factors and cyclooxygenase-2 in prostate cancer. Transl Res 2016;175:116-128. https://doi.org/10.1016/j.trsl.2016.04.002 |
||||
| 55 Kang MJ, Ryu BK, Lee MG, Han J, Lee JH, Ha TK, Byun DS, Chae KS, Lee BH, Chun HS, Lee KY, Kim HJ, Chi SG: NF-kappaB activates transcription of the RNA-binding factor HuR, via PI3K-AKT signaling, to promote gastric tumorigenesis. Gastroenterology 2008;135:2030-2042, 2042e.1-3. https://doi.org/10.1053/j.gastro.2008.08.009 |
||||
| 56 Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, Zhao G: METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer 2019;18:142. https://doi.org/10.1186/s12943-019-1065-4 |
||||
| 57 Wang W, Yang X, Cristofalo VJ, Holbrook NJ, Gorospe M: Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol Cell Biol 2001;21:5889-5898. https://doi.org/10.1128/MCB.21.17.5889-5898.2001 |
||||
| 58 Guo X, Connick MC, Vanderhoof J, Ishak MA, Hartley RS: MicroRNA-16 modulates HuR regulation of cyclin E1 in breast cancer cells. Int J Mol Sci 2015;16:7112-7132. https://doi.org/10.3390/ijms16047112 |
||||
| 59 Yu D, Zhang C, Gui J: RNA-binding protein HuR promotes bladder cancer progression by competitively binding to the long noncoding HOTAIR with miR-1. Onco Targets Ther 2017;10:2609-2619. https://doi.org/10.2147/OTT.S132728 |
||||
| 60 Xu H, Ma J, Zheng J, Wu J, Qu C, Sun F, Xu S: MiR-31 Functions as a Tumor Suppressor in Lung Adenocarcinoma Mainly by Targeting HuR. Clin Lab 2016;62:711-718. https://doi.org/10.7754/Clin.Lab.2015.150903 |
||||
| 61 Poria DK, Guha A, Nandi I, Ray PS: RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4. Oncogene 2016;35:1703-1715. https://doi.org/10.1038/onc.2015.235 |
||||
| 62 Ning M, Chunhua M, Rong J, Yuan L, Jinduo L, Bin W, Liwei S: Diagnostic value of circulating tumor cells in cerebrospinal fluid. Open Med (Wars) 2016;11:21-24. https://doi.org/10.1515/med-2016-0005 |
||||
| 63 Wu T, Shi JX, Geng S, Zhou W, Shi Y, Su X: The MK2/HuR signaling pathway regulates TNF-alpha-induced ICAM-1 expression by promoting the stabilization of ICAM-1 mRNA. BMC Pulm Med 2016;16:84. https://doi.org/10.1186/s12890-016-0247-8 |
||||
| 64 Liu Y, Chen X, Cheng R, Yang F, Yu M, Wang C, Cui S, Hong Y, Liang H, Liu M, Zhao C, Ding M, Sun W, Liu Z, Sun F, Zhang C, Zhou Z, Jiang X, Chen X: The Jun/miR-22/HuR regulatory axis contributes to tumourigenesis in colorectal cancer. Mol Cancer 2018;17:11. https://doi.org/10.1186/s12943-017-0751-3 |
||||